Gene
KWMTBOMO09335
Pre Gene Modal
BGIBMGA002173
Annotation
PREDICTED:_neuroligin-4?_X-linked-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.452
Sequence
CDS
ATGAAGCCTCGGATGAAAAACCATTACCGTGCCCACCAGCTGTCGGTTTGGCTGCGTCTAATCCCGGAAATCCACAGAGCGGGGATGAAGGACGTGGTCGCCAAACACAACCTTTTCAGGAACCACAACGACCCCGAGCTGTACGACGGTTTAGTGCGGCCCGATCCCCTCACCCGGGCAAACTACTTCGATCCGACGCTCGAGCTCTACCGCAAACCGATTTACAACGTAACATTGGACGTTCCGTCCACTACAATGGACACTTACGTCACTACATGTATCAGCGTTATGTCCGCTCGTCCGGGCTCGGCGGTCACACAGAGCCAAGTTCCGAACAACACGCACACTCAGGACGTCTCGAACTTGGAAGTAGCTGGTTACACGGCCTACTCCACAGCGTTAAGTGTTACGATAGCGATCGGTTGTTCCCTACTCATCCTGAACGTTTTAATATTCGCTGGAGTTTATTACCAAAGAGATAAAACGAGATTGCAGGTCAAGGCGTTGCAACAACAGCAAAAGAGGAATCATAATTCGACTTTCGATAGTGTATCTTCAAAGCACCCTCATTATTTTGTGGGACATTCTCAGAGCTCGAGTACGATCGTGGACATCGACCATCAGGACAAAAACGCTATCATAGCCATGACGAATAGGGTGCCCCATTTTACGGGCACAAATTGCCCGAACGTTTGTCACACGGGAATACAAATGTCCAATCTGACGCAAAAATCGAGTCCTCCATCGAATCGAGGTCAATGCACGACATTACCCAGGAAAGTGGGGTTCAGTTACCAACAACCTAATCAGATTTGTAACGCGTCAAACTGCATGACGCTACCTAAGAATGCCACATTTAGTGCTAATAATTTACCCGATGTTCAAGCACAGACTGGACAGTCACAGAGTGCTGGGAACGGTGTACCTCCATCCTCGCCCCAAGCCCAGCACTTCTCCCAAAAGCCAAGAGTACCTCAGGCAGCGATGTCAGAAATGAACGTATGA
Protein
MKPRMKNHYRAHQLSVWLRLIPEIHRAGMKDVVAKHNLFRNHNDPELYDGLVRPDPLTRANYFDPTLELYRKPIYNVTLDVPSTTMDTYVTTCISVMSARPGSAVTQSQVPNNTHTQDVSNLEVAGYTAYSTALSVTIAIGCSLLILNVLIFAGVYYQRDKTRLQVKALQQQQKRNHNSTFDSVSSKHPHYFVGHSQSSSTIVDIDHQDKNAIIAMTNRVPHFTGTNCPNVCHTGIQMSNLTQKSSPPSNRGQCTTLPRKVGFSYQQPNQICNASNCMTLPKNATFSANNLPDVQAQTGQSQSAGNGVPPSSPQAQHFSQKPRVPQAAMSEMNV
Summary
Uniprot
H9IY40
A0A2A4JXL5
A0A2W1BDC8
A0A2H1VKL6
A0A0L7KYL0
A0A3S2NY75
+ More
A0A194Q2A5 A0A194RMK8 A0A212EHX5 A0A1L8D6N7 A0A139WI26 A0A1Y1LPL9 N6THB6 U4TYL9 A0A1B0GLI3 A0A2J7RSF3 A0A1B0D658 A0A1B0DKM2 A0A0L7KMX6 A0A182MF31 A0A067R6L6 B0WXM8 A0A182SWC4 A0A3F2YXM8 A0A182XVM7 A0A182W0E9 A0A182FAB3 A0A182V4R0 A0A182X8E9 A0A182HJ05 A0A182P8D7 A0A182Q539 A0A182N6P2 W5JRC0 Q7PGX1 A0A182JEW4 A0A2H1V8H5 A0A0N0PAM5 A0A0U2LHT4 A0A212FJJ8 A0A437BB17 A0A2A4K584 A0A1U9X1Z5 A0A2W1BEL2 H9IY47 A0A336MYL7 A0A194Q6W9 A0A1J1IDG4 B4PP76 B4LXI9 A0A0M5J2B0 Q9VDP5 B6IDZ4 W8BSI7 B5DVH8 B4NGA7 A0A3B0KJP9 A0A0P8XXC1 B4K9C7 A0A0Q9X167 A0A0Q9X8Z0 A0A023F3M4 A0A1W4VXD5 A0A336M1U8
A0A194Q2A5 A0A194RMK8 A0A212EHX5 A0A1L8D6N7 A0A139WI26 A0A1Y1LPL9 N6THB6 U4TYL9 A0A1B0GLI3 A0A2J7RSF3 A0A1B0D658 A0A1B0DKM2 A0A0L7KMX6 A0A182MF31 A0A067R6L6 B0WXM8 A0A182SWC4 A0A3F2YXM8 A0A182XVM7 A0A182W0E9 A0A182FAB3 A0A182V4R0 A0A182X8E9 A0A182HJ05 A0A182P8D7 A0A182Q539 A0A182N6P2 W5JRC0 Q7PGX1 A0A182JEW4 A0A2H1V8H5 A0A0N0PAM5 A0A0U2LHT4 A0A212FJJ8 A0A437BB17 A0A2A4K584 A0A1U9X1Z5 A0A2W1BEL2 H9IY47 A0A336MYL7 A0A194Q6W9 A0A1J1IDG4 B4PP76 B4LXI9 A0A0M5J2B0 Q9VDP5 B6IDZ4 W8BSI7 B5DVH8 B4NGA7 A0A3B0KJP9 A0A0P8XXC1 B4K9C7 A0A0Q9X167 A0A0Q9X8Z0 A0A023F3M4 A0A1W4VXD5 A0A336M1U8
Pubmed
19121390
28756777
26227816
26354079
22118469
18362917
+ More
19820115 28004739 23537049 24845553 25244985 20920257 23761445 12364791 14747013 17210077 26749290 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 18057021 25474469
19820115 28004739 23537049 24845553 25244985 20920257 23761445 12364791 14747013 17210077 26749290 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 18057021 25474469
EMBL
BABH01029677
NWSH01000461
PCG76243.1
KZ150139
PZC72958.1
ODYU01003066
+ More
SOQ41368.1 JTDY01004463 KOB68129.1 RSAL01000099 RVE47592.1 KQ459581 KPI99129.1 KQ460154 KPJ17231.1 AGBW02014763 OWR41088.1 GEYN01000074 JAV02055.1 KQ971342 KYB27544.1 GEZM01050443 JAV75589.1 APGK01037780 KB740949 ENN77153.1 KB631852 ERL86729.1 AJWK01035053 NEVH01000259 PNF43765.1 AJVK01025765 AJVK01025766 AJVK01067939 JTDY01008721 KOB64431.1 AXCM01014898 KK852665 KDR18954.1 DS232169 EDS36558.1 APCN01002110 AXCN02000220 ADMH02000570 ETN65853.1 AAAB01008879 EAA44773.5 ODYU01001227 SOQ37150.1 KQ461187 KPJ07360.1 KP308206 ALH43424.1 AGBW02008258 OWR53904.1 RVE47584.1 NWSH01000115 PCG79397.1 KY021889 AQY62778.1 KZ150096 PZC73599.1 BABH01029721 BABH01029722 BABH01029723 BABH01029724 UFQT01002602 SSX33753.1 KPI99135.1 CVRI01000047 CRK98256.1 CM000160 EDW96115.2 CH940650 EDW66773.2 CP012526 ALC46886.1 AE014297 AAF55745.4 ACZ94957.1 AHN57419.1 BT050584 ACJ23468.1 ACZ94958.1 GAMC01006682 GAMC01006681 JAB99873.1 CM000070 EDY68564.3 CH964251 EDW83324.2 OUUW01000008 SPP83978.1 CH902617 KPU79345.1 CH933806 EDW15559.2 KRG01621.1 KRG01622.1 KRG01620.1 GBBI01003238 JAC15474.1 UFQT01000426 SSX24215.1
SOQ41368.1 JTDY01004463 KOB68129.1 RSAL01000099 RVE47592.1 KQ459581 KPI99129.1 KQ460154 KPJ17231.1 AGBW02014763 OWR41088.1 GEYN01000074 JAV02055.1 KQ971342 KYB27544.1 GEZM01050443 JAV75589.1 APGK01037780 KB740949 ENN77153.1 KB631852 ERL86729.1 AJWK01035053 NEVH01000259 PNF43765.1 AJVK01025765 AJVK01025766 AJVK01067939 JTDY01008721 KOB64431.1 AXCM01014898 KK852665 KDR18954.1 DS232169 EDS36558.1 APCN01002110 AXCN02000220 ADMH02000570 ETN65853.1 AAAB01008879 EAA44773.5 ODYU01001227 SOQ37150.1 KQ461187 KPJ07360.1 KP308206 ALH43424.1 AGBW02008258 OWR53904.1 RVE47584.1 NWSH01000115 PCG79397.1 KY021889 AQY62778.1 KZ150096 PZC73599.1 BABH01029721 BABH01029722 BABH01029723 BABH01029724 UFQT01002602 SSX33753.1 KPI99135.1 CVRI01000047 CRK98256.1 CM000160 EDW96115.2 CH940650 EDW66773.2 CP012526 ALC46886.1 AE014297 AAF55745.4 ACZ94957.1 AHN57419.1 BT050584 ACJ23468.1 ACZ94958.1 GAMC01006682 GAMC01006681 JAB99873.1 CM000070 EDY68564.3 CH964251 EDW83324.2 OUUW01000008 SPP83978.1 CH902617 KPU79345.1 CH933806 EDW15559.2 KRG01621.1 KRG01622.1 KRG01620.1 GBBI01003238 JAC15474.1 UFQT01000426 SSX24215.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053268
UP000053240
+ More
UP000007151 UP000007266 UP000019118 UP000030742 UP000092461 UP000235965 UP000092462 UP000075883 UP000027135 UP000002320 UP000075901 UP000075900 UP000076408 UP000075920 UP000069272 UP000075903 UP000076407 UP000075840 UP000075885 UP000075886 UP000075884 UP000000673 UP000007062 UP000075880 UP000183832 UP000002282 UP000008792 UP000092553 UP000000803 UP000001819 UP000007798 UP000268350 UP000007801 UP000009192 UP000192221
UP000007151 UP000007266 UP000019118 UP000030742 UP000092461 UP000235965 UP000092462 UP000075883 UP000027135 UP000002320 UP000075901 UP000075900 UP000076408 UP000075920 UP000069272 UP000075903 UP000076407 UP000075840 UP000075885 UP000075886 UP000075884 UP000000673 UP000007062 UP000075880 UP000183832 UP000002282 UP000008792 UP000092553 UP000000803 UP000001819 UP000007798 UP000268350 UP000007801 UP000009192 UP000192221
Pfam
PF00135 COesterase
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9IY40
A0A2A4JXL5
A0A2W1BDC8
A0A2H1VKL6
A0A0L7KYL0
A0A3S2NY75
+ More
A0A194Q2A5 A0A194RMK8 A0A212EHX5 A0A1L8D6N7 A0A139WI26 A0A1Y1LPL9 N6THB6 U4TYL9 A0A1B0GLI3 A0A2J7RSF3 A0A1B0D658 A0A1B0DKM2 A0A0L7KMX6 A0A182MF31 A0A067R6L6 B0WXM8 A0A182SWC4 A0A3F2YXM8 A0A182XVM7 A0A182W0E9 A0A182FAB3 A0A182V4R0 A0A182X8E9 A0A182HJ05 A0A182P8D7 A0A182Q539 A0A182N6P2 W5JRC0 Q7PGX1 A0A182JEW4 A0A2H1V8H5 A0A0N0PAM5 A0A0U2LHT4 A0A212FJJ8 A0A437BB17 A0A2A4K584 A0A1U9X1Z5 A0A2W1BEL2 H9IY47 A0A336MYL7 A0A194Q6W9 A0A1J1IDG4 B4PP76 B4LXI9 A0A0M5J2B0 Q9VDP5 B6IDZ4 W8BSI7 B5DVH8 B4NGA7 A0A3B0KJP9 A0A0P8XXC1 B4K9C7 A0A0Q9X167 A0A0Q9X8Z0 A0A023F3M4 A0A1W4VXD5 A0A336M1U8
A0A194Q2A5 A0A194RMK8 A0A212EHX5 A0A1L8D6N7 A0A139WI26 A0A1Y1LPL9 N6THB6 U4TYL9 A0A1B0GLI3 A0A2J7RSF3 A0A1B0D658 A0A1B0DKM2 A0A0L7KMX6 A0A182MF31 A0A067R6L6 B0WXM8 A0A182SWC4 A0A3F2YXM8 A0A182XVM7 A0A182W0E9 A0A182FAB3 A0A182V4R0 A0A182X8E9 A0A182HJ05 A0A182P8D7 A0A182Q539 A0A182N6P2 W5JRC0 Q7PGX1 A0A182JEW4 A0A2H1V8H5 A0A0N0PAM5 A0A0U2LHT4 A0A212FJJ8 A0A437BB17 A0A2A4K584 A0A1U9X1Z5 A0A2W1BEL2 H9IY47 A0A336MYL7 A0A194Q6W9 A0A1J1IDG4 B4PP76 B4LXI9 A0A0M5J2B0 Q9VDP5 B6IDZ4 W8BSI7 B5DVH8 B4NGA7 A0A3B0KJP9 A0A0P8XXC1 B4K9C7 A0A0Q9X167 A0A0Q9X8Z0 A0A023F3M4 A0A1W4VXD5 A0A336M1U8
PDB
5OJ6
E-value=0.00470582,
Score=93
Ontologies
GO
Topology
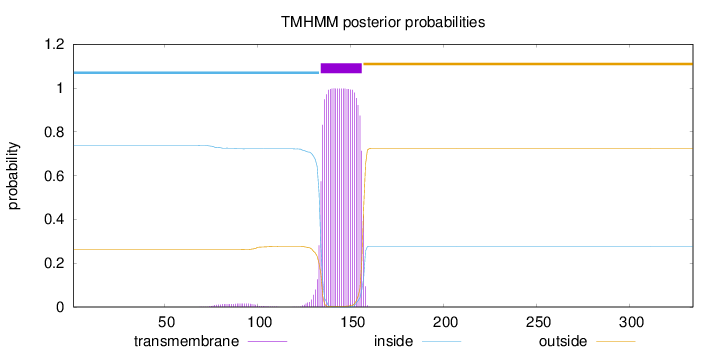
Length:
334
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.24184
Exp number, first 60 AAs:
0.00176
Total prob of N-in:
0.73862
inside
1 - 133
TMhelix
134 - 156
outside
157 - 334
Population Genetic Test Statistics
Pi
247.712857
Theta
232.181894
Tajima's D
0.11233
CLR
0.65394
CSRT
0.40397980100995
Interpretation
Uncertain
