Gene
KWMTBOMO09318
Annotation
PREDICTED:_uncharacterized_protein_LOC105842439_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.598
Sequence
CDS
ATGCAGAGGCTGTTGGACCTACTGCCCCAGTGGCTGGACCGATGGAGGGTCGCGGTCAACGTGGGGAAGACCGCGGCTATCCTCTTCGGTCCGGTCCGCACAAGAGCCGTCCCAGGACAGCTCAGTCTCCGTGGAGTCAATGTCGAGTTTAGGCCCAGTGTCCGATACCTGGGAGTCAATATCGACCGGAGTTTGAGGATGACCGCCCACGCCAAGTCGGCGTTGGCGGCCGCCCGCTATGCGAGGTTTCTCCTCCGGCCGGTGTTGGCCTCCAGGTTGCCGACCAGGACTAGACTCGGCATTTACAAGACGTATGTTCGTTCCCGTATCACGTACGCAGCTGCGGCCTGGTATCACCTCATCCCGCAGATCATGCGGGTGAAGTTACAGGCCCAGCAGAATCTGGCTCTTCGCACGATAGTCGGAGCAGGGCGTTACGTCAGAAATGACGTAATCGCCCGGGATCTAGATGTGGAGTCGCTCGAGGAGTTCATCCGGAGGCTAGCTCGTAATATGTACGAGCGGGCTGACGGTGGACCCCACGAGCATCTCCACAATATAGCTCCCTTGCACGCCAGACCACCTGATGGTCAGGCGCTACCAAGGGAGCTACTGGAACCCCCGGCTATGGTAAGATAG
Protein
MQRLLDLLPQWLDRWRVAVNVGKTAAILFGPVRTRAVPGQLSLRGVNVEFRPSVRYLGVNIDRSLRMTAHAKSALAAARYARFLLRPVLASRLPTRTRLGIYKTYVRSRITYAAAAWYHLIPQIMRVKLQAQQNLALRTIVGAGRYVRNDVIARDLDVESLEEFIRRLARNMYERADGGPHEHLHNIAPLHARPPDGQALPRELLEPPAMVR
Summary
Uniprot
A0A212EVJ8
A0A3S2NJ07
J9LF86
A0A437ATB5
X1WKB2
X1X928
+ More
X1XFE3 X1WR20 X1X2B8 N6UKN2 J9KCZ4 X1WNI2 J9LFC9 A0A1Y1KUU7 X1WWQ0 V5GNF3 J9JUG3 J9KPF1 A0A2S2QZC0 J9L5W8 X1WSV5 A0A2S2Q823 X1WZH0 J9L2G8 A0A1B6G5T1 X1WN18 N6T207 A0A087T1W0 X1X092 U4U2B8 X1X2J8 X1XM65 J9L876 J9LC21 X1WLB2 N6U832 X1X2T1 X1WVE4 X1WMU5 X1XAH1 A0A087U2S8 J9KLQ2 X1X0N4 A0A2S2R1B2 X1XGU0 J9LR29 X1WMF9 X1WVA7 U4UQD3 J9KVX5 A0A2S2PR14 X1X1N9 J9KKL3 V5I850 X1WWC4 A0A2S2QTZ8 A0A2S2NJB5 V5GNJ2 X1WL89 A0A0A9Z4E6 J9LKG7 A0A087TG80 J9JYZ4 U4TYL4 X1X076 J9KF99 J9KPF2 X1WXU3 A0A2S2R0N1 A0A0A9YN51 A0A2J7QIS4 A0A087TU45 A0A087TV70 D6X1N2 X1WM88 U4UTK6 A0A1Y1NA23 A0A2S2N7M6 U4UP62 A0A0A9YJ00 A0A023EZE6 X1WWD6 A0A0N1PFV1 A0A0N0PA09
X1XFE3 X1WR20 X1X2B8 N6UKN2 J9KCZ4 X1WNI2 J9LFC9 A0A1Y1KUU7 X1WWQ0 V5GNF3 J9JUG3 J9KPF1 A0A2S2QZC0 J9L5W8 X1WSV5 A0A2S2Q823 X1WZH0 J9L2G8 A0A1B6G5T1 X1WN18 N6T207 A0A087T1W0 X1X092 U4U2B8 X1X2J8 X1XM65 J9L876 J9LC21 X1WLB2 N6U832 X1X2T1 X1WVE4 X1WMU5 X1XAH1 A0A087U2S8 J9KLQ2 X1X0N4 A0A2S2R1B2 X1XGU0 J9LR29 X1WMF9 X1WVA7 U4UQD3 J9KVX5 A0A2S2PR14 X1X1N9 J9KKL3 V5I850 X1WWC4 A0A2S2QTZ8 A0A2S2NJB5 V5GNJ2 X1WL89 A0A0A9Z4E6 J9LKG7 A0A087TG80 J9JYZ4 U4TYL4 X1X076 J9KF99 J9KPF2 X1WXU3 A0A2S2R0N1 A0A0A9YN51 A0A2J7QIS4 A0A087TU45 A0A087TV70 D6X1N2 X1WM88 U4UTK6 A0A1Y1NA23 A0A2S2N7M6 U4UP62 A0A0A9YJ00 A0A023EZE6 X1WWD6 A0A0N1PFV1 A0A0N0PA09
EMBL
AGBW02012184
OWR45487.1
RSAL01000708
RVE41206.1
ABLF02017815
RSAL01000536
+ More
RVE41424.1 ABLF02049496 ABLF02066620 ABLF02029291 ABLF02016515 ABLF02016517 ABLF02041765 APGK01020047 KB740155 ENN81216.1 ABLF02008463 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02023911 GEZM01074951 JAV64378.1 ABLF02021994 GALX01005359 JAB63107.1 ABLF02008677 ABLF02011967 ABLF02052409 ABLF02056408 ABLF02041767 GGMS01013269 MBY82472.1 ABLF02015272 ABLF02016494 ABLF02066976 GGMS01004685 MBY73888.1 ABLF02034467 ABLF02003375 GECZ01011980 JAS57789.1 ABLF02021177 ABLF02027367 ABLF02066338 APGK01055369 APGK01055370 KB741264 ENN71543.1 KK113014 KFM59099.1 ABLF02026276 KB631186 ERL84155.1 ABLF02061876 ABLF02030700 ABLF02033522 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 ABLF02042518 ABLF02057746 APGK01045324 KB741033 ENN74747.1 ABLF02003699 ABLF02003700 ABLF02007662 ABLF02007663 ABLF02019401 ABLF02049974 ABLF02050034 ABLF02004189 ABLF02005775 ABLF02015592 ABLF02057149 ABLF02058897 ABLF02014862 ABLF02014866 ABLF02061908 ABLF02046613 KK117880 KFM71667.1 ABLF02030319 ABLF02030320 ABLF02060395 GGMS01014520 MBY83723.1 ABLF02055645 ABLF02025082 ABLF02039175 ABLF02041809 ABLF02061874 ABLF02034494 KB632420 ERL95312.1 ABLF02010251 ABLF02035981 GGMR01019198 MBY31817.1 ABLF02052197 ABLF02029306 ABLF02029314 ABLF02060138 GALX01005300 JAB63166.1 ABLF02003961 ABLF02059872 GGMS01012005 MBY81208.1 GGMR01004661 MBY17280.1 GALX01005299 JAB63167.1 ABLF02019349 ABLF02041900 GBHO01041571 GBHO01005400 GBHO01005396 JAG02033.1 JAG38204.1 JAG38208.1 ABLF02017810 ABLF02017816 KK115079 KFM64119.1 KB631851 ERL86724.1 ABLF02017659 ABLF02017667 ABLF02065648 ABLF02014933 ABLF02014938 ABLF02003739 ABLF02003740 ABLF02008933 GGMS01014127 MBY83330.1 GBHO01041572 GBHO01041570 GBHO01010553 JAG02032.1 JAG02034.1 JAG33051.1 NEVH01013579 PNF28469.1 KK116737 KFM68634.1 KK116883 KFM69009.1 KQ971370 EFA09320.1 ABLF02062685 KB632361 ERL93526.1 GEZM01008457 JAV94772.1 GGMR01000167 MBY12786.1 KB632400 ERL94892.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 GBBI01004571 JAC14141.1 ABLF02042106 KQ459265 KPJ02064.1 KQ459259 KPJ02068.1
RVE41424.1 ABLF02049496 ABLF02066620 ABLF02029291 ABLF02016515 ABLF02016517 ABLF02041765 APGK01020047 KB740155 ENN81216.1 ABLF02008463 ABLF02008153 ABLF02008462 ABLF02008464 ABLF02023911 GEZM01074951 JAV64378.1 ABLF02021994 GALX01005359 JAB63107.1 ABLF02008677 ABLF02011967 ABLF02052409 ABLF02056408 ABLF02041767 GGMS01013269 MBY82472.1 ABLF02015272 ABLF02016494 ABLF02066976 GGMS01004685 MBY73888.1 ABLF02034467 ABLF02003375 GECZ01011980 JAS57789.1 ABLF02021177 ABLF02027367 ABLF02066338 APGK01055369 APGK01055370 KB741264 ENN71543.1 KK113014 KFM59099.1 ABLF02026276 KB631186 ERL84155.1 ABLF02061876 ABLF02030700 ABLF02033522 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 ABLF02042518 ABLF02057746 APGK01045324 KB741033 ENN74747.1 ABLF02003699 ABLF02003700 ABLF02007662 ABLF02007663 ABLF02019401 ABLF02049974 ABLF02050034 ABLF02004189 ABLF02005775 ABLF02015592 ABLF02057149 ABLF02058897 ABLF02014862 ABLF02014866 ABLF02061908 ABLF02046613 KK117880 KFM71667.1 ABLF02030319 ABLF02030320 ABLF02060395 GGMS01014520 MBY83723.1 ABLF02055645 ABLF02025082 ABLF02039175 ABLF02041809 ABLF02061874 ABLF02034494 KB632420 ERL95312.1 ABLF02010251 ABLF02035981 GGMR01019198 MBY31817.1 ABLF02052197 ABLF02029306 ABLF02029314 ABLF02060138 GALX01005300 JAB63166.1 ABLF02003961 ABLF02059872 GGMS01012005 MBY81208.1 GGMR01004661 MBY17280.1 GALX01005299 JAB63167.1 ABLF02019349 ABLF02041900 GBHO01041571 GBHO01005400 GBHO01005396 JAG02033.1 JAG38204.1 JAG38208.1 ABLF02017810 ABLF02017816 KK115079 KFM64119.1 KB631851 ERL86724.1 ABLF02017659 ABLF02017667 ABLF02065648 ABLF02014933 ABLF02014938 ABLF02003739 ABLF02003740 ABLF02008933 GGMS01014127 MBY83330.1 GBHO01041572 GBHO01041570 GBHO01010553 JAG02032.1 JAG02034.1 JAG33051.1 NEVH01013579 PNF28469.1 KK116737 KFM68634.1 KK116883 KFM69009.1 KQ971370 EFA09320.1 ABLF02062685 KB632361 ERL93526.1 GEZM01008457 JAV94772.1 GGMR01000167 MBY12786.1 KB632400 ERL94892.1 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 GBBI01004571 JAC14141.1 ABLF02042106 KQ459265 KPJ02064.1 KQ459259 KPJ02068.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A212EVJ8
A0A3S2NJ07
J9LF86
A0A437ATB5
X1WKB2
X1X928
+ More
X1XFE3 X1WR20 X1X2B8 N6UKN2 J9KCZ4 X1WNI2 J9LFC9 A0A1Y1KUU7 X1WWQ0 V5GNF3 J9JUG3 J9KPF1 A0A2S2QZC0 J9L5W8 X1WSV5 A0A2S2Q823 X1WZH0 J9L2G8 A0A1B6G5T1 X1WN18 N6T207 A0A087T1W0 X1X092 U4U2B8 X1X2J8 X1XM65 J9L876 J9LC21 X1WLB2 N6U832 X1X2T1 X1WVE4 X1WMU5 X1XAH1 A0A087U2S8 J9KLQ2 X1X0N4 A0A2S2R1B2 X1XGU0 J9LR29 X1WMF9 X1WVA7 U4UQD3 J9KVX5 A0A2S2PR14 X1X1N9 J9KKL3 V5I850 X1WWC4 A0A2S2QTZ8 A0A2S2NJB5 V5GNJ2 X1WL89 A0A0A9Z4E6 J9LKG7 A0A087TG80 J9JYZ4 U4TYL4 X1X076 J9KF99 J9KPF2 X1WXU3 A0A2S2R0N1 A0A0A9YN51 A0A2J7QIS4 A0A087TU45 A0A087TV70 D6X1N2 X1WM88 U4UTK6 A0A1Y1NA23 A0A2S2N7M6 U4UP62 A0A0A9YJ00 A0A023EZE6 X1WWD6 A0A0N1PFV1 A0A0N0PA09
X1XFE3 X1WR20 X1X2B8 N6UKN2 J9KCZ4 X1WNI2 J9LFC9 A0A1Y1KUU7 X1WWQ0 V5GNF3 J9JUG3 J9KPF1 A0A2S2QZC0 J9L5W8 X1WSV5 A0A2S2Q823 X1WZH0 J9L2G8 A0A1B6G5T1 X1WN18 N6T207 A0A087T1W0 X1X092 U4U2B8 X1X2J8 X1XM65 J9L876 J9LC21 X1WLB2 N6U832 X1X2T1 X1WVE4 X1WMU5 X1XAH1 A0A087U2S8 J9KLQ2 X1X0N4 A0A2S2R1B2 X1XGU0 J9LR29 X1WMF9 X1WVA7 U4UQD3 J9KVX5 A0A2S2PR14 X1X1N9 J9KKL3 V5I850 X1WWC4 A0A2S2QTZ8 A0A2S2NJB5 V5GNJ2 X1WL89 A0A0A9Z4E6 J9LKG7 A0A087TG80 J9JYZ4 U4TYL4 X1X076 J9KF99 J9KPF2 X1WXU3 A0A2S2R0N1 A0A0A9YN51 A0A2J7QIS4 A0A087TU45 A0A087TV70 D6X1N2 X1WM88 U4UTK6 A0A1Y1NA23 A0A2S2N7M6 U4UP62 A0A0A9YJ00 A0A023EZE6 X1WWD6 A0A0N1PFV1 A0A0N0PA09
Ontologies
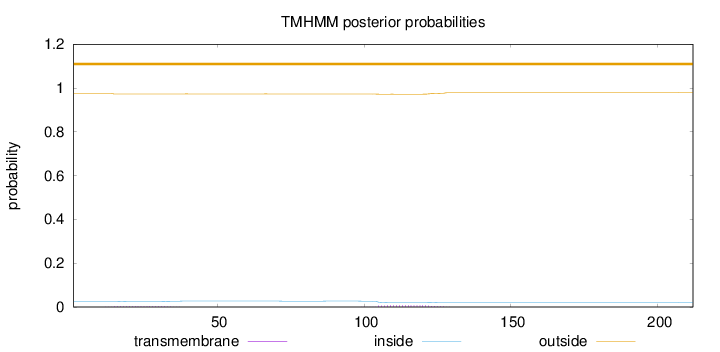
Topology
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21305
Exp number, first 60 AAs:
0.05743
Total prob of N-in:
0.02506
outside
1 - 212
Population Genetic Test Statistics
Pi
369.678885
Theta
199.629192
Tajima's D
2.609385
CLR
0
CSRT
0.952952352382381
Interpretation
Uncertain
