Gene
KWMTBOMO09304
Pre Gene Modal
BGIBMGA003307
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_RNF13_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.285
Sequence
CDS
ATGGAGGAATTCCGCGACATCCCGGCTAGTTTCGGCGAGGAACTGCCGCCGGAGGGTCTGCGCGGACTCCTGGTCGAGGGCGTCCCGGCTGACGGATGCACGGCCCTGGATAAACCGCCCGTTGTCGACAGCTTTACCGGGAAATGGATCGTACTCGTAGCCAGATACAACTGCAGCTTCGAAGTGAAGATACGCAACGCGCAGGCGGCCGGCTTCGATTGCGCGATCGTGCACAACGTGAACTCGAGCGATTTGGAGAAAATGTCGGCTAAGAATCCGGAGGGCATCGTGATACCTTCAGTGTTCGTCAGTGACGTGGCCGGCGACATACTGGCAGCGGAATACCAATATAATGCCGGTTTCTTTATAATGGTGAACGGCGACCTACCGTTCAACATCAACACGCACCTGCTGTTGCCCTTCGCAATAGTGGTCGTGCTCTGTCTACTCGTCATATTTATATTTATGATAGTGAAATGTATAAAAGACAGACGCAGAGCCAGAAGGCACCGCTTACCGCATCGTGCGCTTTCGAAGATACCGACGTGCAAGTTCAGCAAGGGCGACCCGTACGACACGTGCGCCATCTGCCTTGACGACTACCAGGAGGGGGACCGCCTGCGCGTGCTGCCCTGCGCCCACGCTTACCACAGCGCGTGCATCGACCCGTGGCTGACGCAGAACAAGCGCGTGTGTCCGGTGTGCAAGCGCCGCGTGGTGTCCGCCGGCGAGCCCCGCCCCCGCCGCTCCTCCTCCTCCTCCGACTCCGACACCGAGCCGCTCCTGCCGCAGCCGCGGGACCAGGGAGGAGGAACATTTGAAGAGCAACGCGAGAATCCTTTCGTGCGAGCGGCGAGATTCATGTCGAGGTTGCGGGCTCGAAATAACCAGAATAGTGCTGATGCAGAGGTCCCTCCTGAACTAGAAGATAGTCAAGAAACGTCTGATCAGGACGAATTGAACGGCAGTACCGAGGCGCAGCCCCTGCTGGAGGAGCGCGCCCCCCACTCCATCAACGGCTCCTCGCGCGGGGAGATCGGGGTGGCGGCGCTGCCGGCTCCCGCCTACCACTCGCTGCCGCGGTACATCGACTTATGA
Protein
MEEFRDIPASFGEELPPEGLRGLLVEGVPADGCTALDKPPVVDSFTGKWIVLVARYNCSFEVKIRNAQAAGFDCAIVHNVNSSDLEKMSAKNPEGIVIPSVFVSDVAGDILAAEYQYNAGFFIMVNGDLPFNINTHLLLPFAIVVVLCLLVIFIFMIVKCIKDRRRARRHRLPHRALSKIPTCKFSKGDPYDTCAICLDDYQEGDRLRVLPCAHAYHSACIDPWLTQNKRVCPVCKRRVVSAGEPRPRRSSSSSDSDTEPLLPQPRDQGGGTFEEQRENPFVRAARFMSRLRARNNQNSADAEVPPELEDSQETSDQDELNGSTEAQPLLEERAPHSINGSSRGEIGVAALPAPAYHSLPRYIDL
Summary
Uniprot
A0A1E1WVK6
A0A3S2NIR9
A0A1E1WQF6
A0A194RKH0
A0A2A4JQ86
A0A2W1BWU1
+ More
A0A0C9QY57 A0A0J7NEF8 A0A026WDU9 E1ZWF2 A0A2A3EAX2 A0A1W4WUL7 V9IIJ1 A0A195EW65 F4WV10 A0A151J1N6 E9J9L8 A0A088AHH3 A0A1W4WUB3 A0A151WT35 A0A195BS18 A0A158NNT3 A0A2J7PE15 A0A2J7PE22 A0A151I7N6 A0A310S6D2 A0A0M8ZR78 A0A154NZR8 D7EL38 A0A232FEL3 A0A1B6M4I7 A0A1Y1LIE5 A0A212FEQ1 A0A224XNQ8 A0A0V0GAY0 T1HBC2 N6UAC4 E9GUD5 A0A0N8AZC6 A0A0P5VCK9 A0A0P5MYQ8 A0A0P5IU71 A0A0P5TJ08 A0A0P5RE42 A0A0N8EP78 A0A0P5EC08 A0A0P5FIL4
A0A0C9QY57 A0A0J7NEF8 A0A026WDU9 E1ZWF2 A0A2A3EAX2 A0A1W4WUL7 V9IIJ1 A0A195EW65 F4WV10 A0A151J1N6 E9J9L8 A0A088AHH3 A0A1W4WUB3 A0A151WT35 A0A195BS18 A0A158NNT3 A0A2J7PE15 A0A2J7PE22 A0A151I7N6 A0A310S6D2 A0A0M8ZR78 A0A154NZR8 D7EL38 A0A232FEL3 A0A1B6M4I7 A0A1Y1LIE5 A0A212FEQ1 A0A224XNQ8 A0A0V0GAY0 T1HBC2 N6UAC4 E9GUD5 A0A0N8AZC6 A0A0P5VCK9 A0A0P5MYQ8 A0A0P5IU71 A0A0P5TJ08 A0A0P5RE42 A0A0N8EP78 A0A0P5EC08 A0A0P5FIL4
Pubmed
EMBL
GDQN01006708
GDQN01004963
GDQN01000040
JAT84346.1
JAT86091.1
JAT91014.1
+ More
RSAL01000026 RVE52074.1 GDQN01001907 JAT89147.1 KQ460106 KPJ17830.1 NWSH01000846 PCG73868.1 KZ149923 PZC77667.1 GBYB01001634 GBYB01005617 GBYB01005618 GBYB01005619 JAG71401.1 JAG75384.1 JAG75385.1 JAG75386.1 LBMM01006115 KMQ90915.1 KK107256 QOIP01000006 EZA54275.1 RLU22165.1 GL434819 EFN74498.1 KZ288310 PBC28624.1 JR046038 AEY60119.1 KQ981953 KYN32478.1 GL888384 EGI61898.1 KQ980503 KYN15789.1 GL769399 EFZ10438.1 KQ982766 KYQ50993.1 KQ976417 KYM89810.1 ADTU01021753 ADTU01021754 NEVH01026112 PNF14580.1 PNF14577.1 KQ978438 KYM93977.1 KQ794714 OAD47015.1 KQ435969 KOX67879.1 KQ434786 KZC05093.1 DS497802 EFA11839.1 NNAY01000354 OXU28960.1 GEBQ01009159 JAT30818.1 GEZM01060990 GEZM01060989 JAV70797.1 AGBW02008909 OWR52221.1 GFTR01006356 JAW10070.1 GECL01000897 JAP05227.1 ACPB03005610 APGK01032712 KB740848 KB632411 ENN78680.1 ERL95235.1 GL732566 EFX76866.1 GDIQ01228051 JAK23674.1 GDIP01117037 JAL86677.1 GDIQ01149888 LRGB01000115 JAL01838.1 KZS20779.1 GDIQ01210462 JAK41263.1 GDIP01129085 JAL74629.1 GDIQ01102702 JAL49024.1 GDIQ01007444 JAN87293.1 GDIP01145085 JAJ78317.1 GDIP01147699 GDIQ01184315 JAJ75703.1 JAK67410.1
RSAL01000026 RVE52074.1 GDQN01001907 JAT89147.1 KQ460106 KPJ17830.1 NWSH01000846 PCG73868.1 KZ149923 PZC77667.1 GBYB01001634 GBYB01005617 GBYB01005618 GBYB01005619 JAG71401.1 JAG75384.1 JAG75385.1 JAG75386.1 LBMM01006115 KMQ90915.1 KK107256 QOIP01000006 EZA54275.1 RLU22165.1 GL434819 EFN74498.1 KZ288310 PBC28624.1 JR046038 AEY60119.1 KQ981953 KYN32478.1 GL888384 EGI61898.1 KQ980503 KYN15789.1 GL769399 EFZ10438.1 KQ982766 KYQ50993.1 KQ976417 KYM89810.1 ADTU01021753 ADTU01021754 NEVH01026112 PNF14580.1 PNF14577.1 KQ978438 KYM93977.1 KQ794714 OAD47015.1 KQ435969 KOX67879.1 KQ434786 KZC05093.1 DS497802 EFA11839.1 NNAY01000354 OXU28960.1 GEBQ01009159 JAT30818.1 GEZM01060990 GEZM01060989 JAV70797.1 AGBW02008909 OWR52221.1 GFTR01006356 JAW10070.1 GECL01000897 JAP05227.1 ACPB03005610 APGK01032712 KB740848 KB632411 ENN78680.1 ERL95235.1 GL732566 EFX76866.1 GDIQ01228051 JAK23674.1 GDIP01117037 JAL86677.1 GDIQ01149888 LRGB01000115 JAL01838.1 KZS20779.1 GDIQ01210462 JAK41263.1 GDIP01129085 JAL74629.1 GDIQ01102702 JAL49024.1 GDIQ01007444 JAN87293.1 GDIP01145085 JAJ78317.1 GDIP01147699 GDIQ01184315 JAJ75703.1 JAK67410.1
Proteomes
UP000283053
UP000053240
UP000218220
UP000036403
UP000053097
UP000279307
+ More
UP000000311 UP000242457 UP000192223 UP000078541 UP000007755 UP000078492 UP000005203 UP000075809 UP000078540 UP000005205 UP000235965 UP000078542 UP000053105 UP000076502 UP000007266 UP000215335 UP000007151 UP000015103 UP000019118 UP000030742 UP000000305 UP000076858
UP000000311 UP000242457 UP000192223 UP000078541 UP000007755 UP000078492 UP000005203 UP000075809 UP000078540 UP000005205 UP000235965 UP000078542 UP000053105 UP000076502 UP000007266 UP000215335 UP000007151 UP000015103 UP000019118 UP000030742 UP000000305 UP000076858
Gene 3D
ProteinModelPortal
A0A1E1WVK6
A0A3S2NIR9
A0A1E1WQF6
A0A194RKH0
A0A2A4JQ86
A0A2W1BWU1
+ More
A0A0C9QY57 A0A0J7NEF8 A0A026WDU9 E1ZWF2 A0A2A3EAX2 A0A1W4WUL7 V9IIJ1 A0A195EW65 F4WV10 A0A151J1N6 E9J9L8 A0A088AHH3 A0A1W4WUB3 A0A151WT35 A0A195BS18 A0A158NNT3 A0A2J7PE15 A0A2J7PE22 A0A151I7N6 A0A310S6D2 A0A0M8ZR78 A0A154NZR8 D7EL38 A0A232FEL3 A0A1B6M4I7 A0A1Y1LIE5 A0A212FEQ1 A0A224XNQ8 A0A0V0GAY0 T1HBC2 N6UAC4 E9GUD5 A0A0N8AZC6 A0A0P5VCK9 A0A0P5MYQ8 A0A0P5IU71 A0A0P5TJ08 A0A0P5RE42 A0A0N8EP78 A0A0P5EC08 A0A0P5FIL4
A0A0C9QY57 A0A0J7NEF8 A0A026WDU9 E1ZWF2 A0A2A3EAX2 A0A1W4WUL7 V9IIJ1 A0A195EW65 F4WV10 A0A151J1N6 E9J9L8 A0A088AHH3 A0A1W4WUB3 A0A151WT35 A0A195BS18 A0A158NNT3 A0A2J7PE15 A0A2J7PE22 A0A151I7N6 A0A310S6D2 A0A0M8ZR78 A0A154NZR8 D7EL38 A0A232FEL3 A0A1B6M4I7 A0A1Y1LIE5 A0A212FEQ1 A0A224XNQ8 A0A0V0GAY0 T1HBC2 N6UAC4 E9GUD5 A0A0N8AZC6 A0A0P5VCK9 A0A0P5MYQ8 A0A0P5IU71 A0A0P5TJ08 A0A0P5RE42 A0A0N8EP78 A0A0P5EC08 A0A0P5FIL4
PDB
5ZC4
E-value=1.53414e-17,
Score=219
Ontologies
GO
Topology
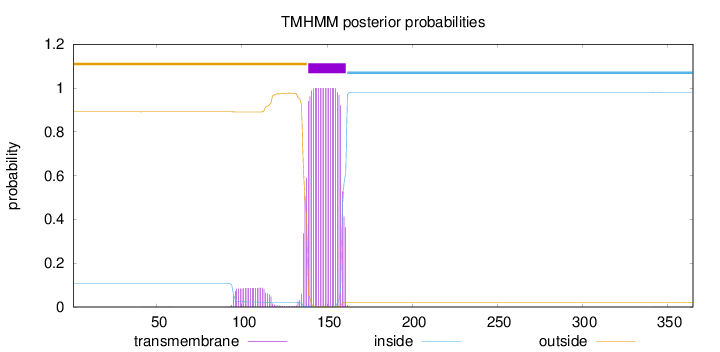
Length:
365
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.53841
Exp number, first 60 AAs:
0.00402
Total prob of N-in:
0.10853
outside
1 - 138
TMhelix
139 - 161
inside
162 - 365
Population Genetic Test Statistics
Pi
182.540418
Theta
156.454854
Tajima's D
0.468913
CLR
0.799117
CSRT
0.511424428778561
Interpretation
Uncertain
