Gene
KWMTBOMO09300 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003309
Annotation
ribosomal_protein_P0_[Bombyx_mori]
Full name
60S acidic ribosomal protein P0
Alternative Name
Apurinic-apyrimidinic endonuclease
DNA-(apurinic or apyrimidinic site) lyase
DNA-(apurinic or apyrimidinic site) lyase
Location in the cell
Cytoplasmic Reliability : 2.335
Sequence
CDS
ATGGGTAGGGAGGACAAGGCTACTTGGAAGTCTAACTACTTCGTTAAGATCATCCAACTCTTGGACGAGTACCCAAAATGTTTCATCGTGGGTGCCGATAACGTGGGCTCGCAACAGATGCAGCAGATCCGTATCTCGCTACGTGGCTCCAGTATCGTGCTCATGGGAAAAAACACAATGATGCGCAAAGCCATCAAAGACCACCTGGACAACAATCCAGCCCTCGAGAAACTGTTGCCACACATCAAGGGCAACGTTGGCTTCGTGTTCACCCGCGGAGACCTCGTTGAGGTCCGTGACAAACTGTTGGAGAACAAAGTCCAAGCTCCAGCTCGTCCTGGTGCCATTGCCCCATTGTCAGTCGTCATTCCCGCCCACAACACCGGCCTCGGTCCAGAGAAGACCTCTTTCTTCCAGGCTCTTTCTATCCCTACCAAGATTTCAAAGGGTACTATTGAAATCATCAACGATGTACACATCTTGAAGCCCGGTGACAAGGTTGGAGCTTCTGAAGCCACCCTTCTCAACATGTTGAACATCTCTCCATTCTCATATGGTCTTGTTGTTAAGCAGGTATATGATTCTGGAACTATTTTTGCACCTGAAATTCTGGACATCAAACCAGAAGATCTCCGTGCCAAGTTCCAAGCTGGAGTTGCTAATGTAGCTGCTCTTTCTTTGGCTATTGGTTACCCAACTATTGCTTCAGCCCCGCATTCCATTGCCAATGGTTTCAAGAACCTTTTGGCCATCGCTGCTGTCACAGAGGTTGAGTTTGAAGAAGCTACCACCATCAAGGAGTTCATTAAGGACCCATCAAAGTTCGCTGCAGTTGCTGCCGCGGTGGCACCGTCCGCCGCCGCTGCTCCGGCTGAGAAGAAGGAGGAAAAGAAAGAAGAAAAAGAGGAGGAAGAAAGCGATGACGACATGGGCTTTGGTCTCTTCGACTAG
Protein
MGREDKATWKSNYFVKIIQLLDEYPKCFIVGADNVGSQQMQQIRISLRGSSIVLMGKNTMMRKAIKDHLDNNPALEKLLPHIKGNVGFVFTRGDLVEVRDKLLENKVQAPARPGAIAPLSVVIPAHNTGLGPEKTSFFQALSIPTKISKGTIEIINDVHILKPGDKVGASEATLLNMLNISPFSYGLVVKQVYDSGTIFAPEILDIKPEDLRAKFQAGVANVAALSLAIGYPTIASAPHSIANGFKNLLAIAAVTEVEFEEATTIKEFIKDPSKFAAVAAAVAPSAAAAPAEKKEEKKEEKEEEESDDDMGFGLFD
Summary
Description
Ribosomal protein P0 is the functional equivalent of E.coli protein L10.
Catalytic Activity
The C-O-P bond 3' to the apurinic or apyrimidinic site in DNA is broken by a beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate.
Subunit
P0 forms a pentameric complex by interaction with dimers of P1 and P2.
Similarity
Belongs to the universal ribosomal protein uL10 family.
Keywords
Phosphoprotein
Ribonucleoprotein
Ribosomal protein
Complete proteome
Cytoplasm
DNA damage
DNA repair
Lyase
Nucleus
Reference proteome
Feature
chain 60S acidic ribosomal protein P0
Uniprot
Q70YZ3
Q5UAU1
A0A212FEQ6
G0ZEC4
E7DZ05
I4DID8
+ More
A0A194RK76 A0A2A4J9D4 A0A0K8SL83 A0A0L7LDY8 D1LYK0 I4DLY9 A0A3G1T1B4 Q8WQJ2 A0A3S2LR49 A0A222AJ53 A0A185RX45 E2BW93 D6WMT4 E2BWC5 A0A348G679 A0A0N0BIW7 Q6EV08 K7IQV7 E9II14 E2AZT8 A0A0C9RCN3 I6P504 V5GRW6 A0A2A3E478 V9IFN9 A0A087ZPA5 A0A0K8TQM2 I6P155 I6P6U6 A0A411G6P8 E6ZCJ9 A0A154NZP4 A0A026VZT8 A0A1B2M4Y0 Q6EV09 A0A0J7KN60 F4X3G3 A0A195BNB3 A0A195FCE2 A0A158NKQ4 A0A151X8W5 B0WQZ4 A0A1Q3FEK8 A0A195CJ37 J3JU33 Q1HR99 A0A195DHA1 J3JV87 A0A023EQJ9 A0A182Q7D3 A0A146LPN5 A0A182LZB4 A0A182K8J7 A0A182RZI1 A0A2M3YX70 A0A182FNS5 A0A2M3YXC4 A0A2M4BUA9 A0A182IRR4 A0A1Y1LK51 A0A2M3ZZC9 W5JMB2 A0A1Y9IV09 A0A0A1WZ39 Q8MQT0 A0A1B0D3Y1 B3MAK8 A0A182KXJ9 A0A182U5N4 A0A182HZC3 A0A182V7K1 A0A182WVR6 Q7PQG7 A0A336MDC9 A0A034VBX9 A0A067R8K2 B4IXN3 A0A109NIB3 W8CB96 Q9U3U0 Q9NHP0 Q29DM5 A0A182N4R2 A0A3B0JLU0 A0A0K8UBE5 A0A1W4VAZ1 M9PG76 P19889 B3NJ27 A0A1A9UP79 B4QKR0 B4IAY7 U5EVH7 A0A0J9RZV4
A0A194RK76 A0A2A4J9D4 A0A0K8SL83 A0A0L7LDY8 D1LYK0 I4DLY9 A0A3G1T1B4 Q8WQJ2 A0A3S2LR49 A0A222AJ53 A0A185RX45 E2BW93 D6WMT4 E2BWC5 A0A348G679 A0A0N0BIW7 Q6EV08 K7IQV7 E9II14 E2AZT8 A0A0C9RCN3 I6P504 V5GRW6 A0A2A3E478 V9IFN9 A0A087ZPA5 A0A0K8TQM2 I6P155 I6P6U6 A0A411G6P8 E6ZCJ9 A0A154NZP4 A0A026VZT8 A0A1B2M4Y0 Q6EV09 A0A0J7KN60 F4X3G3 A0A195BNB3 A0A195FCE2 A0A158NKQ4 A0A151X8W5 B0WQZ4 A0A1Q3FEK8 A0A195CJ37 J3JU33 Q1HR99 A0A195DHA1 J3JV87 A0A023EQJ9 A0A182Q7D3 A0A146LPN5 A0A182LZB4 A0A182K8J7 A0A182RZI1 A0A2M3YX70 A0A182FNS5 A0A2M3YXC4 A0A2M4BUA9 A0A182IRR4 A0A1Y1LK51 A0A2M3ZZC9 W5JMB2 A0A1Y9IV09 A0A0A1WZ39 Q8MQT0 A0A1B0D3Y1 B3MAK8 A0A182KXJ9 A0A182U5N4 A0A182HZC3 A0A182V7K1 A0A182WVR6 Q7PQG7 A0A336MDC9 A0A034VBX9 A0A067R8K2 B4IXN3 A0A109NIB3 W8CB96 Q9U3U0 Q9NHP0 Q29DM5 A0A182N4R2 A0A3B0JLU0 A0A0K8UBE5 A0A1W4VAZ1 M9PG76 P19889 B3NJ27 A0A1A9UP79 B4QKR0 B4IAY7 U5EVH7 A0A0J9RZV4
Pubmed
14527730
19121390
22118469
22651552
26354079
26227816
+ More
14668217 20798317 18362917 19820115 20075255 21282665 23569222 26369729 24508170 30249741 21719571 21347285 22516182 23537049 17204158 17510324 24945155 26483478 26823975 28004739 20920257 23761445 25830018 12903741 17994087 20966253 12364791 14747013 17210077 25348373 24845553 24495485 10672071 11024299 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2471063 12537569 1870984 8932386 18327897 22936249
14668217 20798317 18362917 19820115 20075255 21282665 23569222 26369729 24508170 30249741 21719571 21347285 22516182 23537049 17204158 17510324 24945155 26483478 26823975 28004739 20920257 23761445 25830018 12903741 17994087 20966253 12364791 14747013 17210077 25348373 24845553 24495485 10672071 11024299 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2471063 12537569 1870984 8932386 18327897 22936249
EMBL
AJ457827
CAD29995.1
BABH01023979
AY769267
AAV34809.1
AGBW02008909
+ More
OWR52222.1 JF265002 AEL28833.1 HQ424708 ADT80652.1 AK401056 KQ459602 BAM17678.1 KPI93614.1 KQ460106 KPJ17829.1 NWSH01002527 PCG68070.1 GBRD01011753 JAG54071.1 JTDY01001598 KOB73421.1 GU084274 ACY95307.1 AK402307 BAM18929.1 MG846922 AXY94774.1 AY072284 ODYU01001255 AAL62465.1 SOQ37213.1 RSAL01000026 RVE52072.1 KY564020 ASO76399.1 KM229767 AKC05613.1 GL451103 EFN79997.1 KQ971341 EFA04511.1 EFN80029.1 FX985621 BBF97952.1 KQ435726 KOX78148.1 AJ783863 CAH04311.1 GL763324 EFZ19789.1 GL444277 EFN61067.1 GBYB01006060 JAG75827.1 JQ771366 AFD93397.1 GALX01004089 JAB64377.1 KZ288381 PBC26515.1 JR041657 AEY59487.1 GDAI01001408 JAI16195.1 JQ771358 AFD93389.1 JQ771367 AFD93398.1 MH365708 QBB01517.1 FN908680 CBM69268.1 KQ434786 KZC05146.1 KK107538 QOIP01000005 EZA49150.1 RLU23180.1 KX447597 AOA60264.1 AJ783862 CAH04310.1 LBMM01005253 KMQ91679.1 GL888613 EGI58991.1 KQ976439 KYM86708.1 KQ981693 KYN37872.1 ADTU01018960 KQ982409 KYQ56817.1 DS232047 EDS33081.1 GFDL01009055 JAV25990.1 KQ977649 KYN00740.1 APGK01051867 BT126745 KB741207 KB632327 AEE61707.1 ENN72907.1 ERL92523.1 DQ440195 CH477695 ABF18228.1 EAT37166.1 KQ980886 KYN11839.1 BT127154 AEE62116.1 JXUM01082821 GAPW01002729 KQ563338 JAC10869.1 KXJ74024.1 AXCN02000015 GDHC01008765 JAQ09864.1 AXCM01001019 GGFM01000110 MBW20861.1 GGFM01000150 MBW20901.1 GGFJ01007529 MBW56670.1 GEZM01053565 JAV74013.1 GGFK01000447 MBW33768.1 ADMH02001019 ETN64428.1 GBXI01010522 JAD03770.1 AY127558 AAM97779.1 AJVK01011094 CH902618 EDV41295.1 APCN01005540 AAAB01008888 EAA08855.2 UFQS01000821 UFQT01000410 UFQT01000821 SSX07034.1 SSX24038.1 GAKP01019889 JAC39063.1 KK852865 KDR14762.1 CH916366 EDV97495.1 KC507365 AHB12488.1 GAMC01000654 JAC05902.1 Y18444 AF220939 AAF31449.1 CH379070 EAL30389.1 OUUW01000002 SPP76390.1 GDHF01029382 GDHF01028350 JAI22932.1 JAI23964.1 AE014296 AGB94895.1 M25772 AY075528 BT021447 CH954178 EDV52744.1 CM000363 EDX11475.1 CH480826 EDW44450.1 GANO01001021 JAB58850.1 CM002912 KMZ01149.1
OWR52222.1 JF265002 AEL28833.1 HQ424708 ADT80652.1 AK401056 KQ459602 BAM17678.1 KPI93614.1 KQ460106 KPJ17829.1 NWSH01002527 PCG68070.1 GBRD01011753 JAG54071.1 JTDY01001598 KOB73421.1 GU084274 ACY95307.1 AK402307 BAM18929.1 MG846922 AXY94774.1 AY072284 ODYU01001255 AAL62465.1 SOQ37213.1 RSAL01000026 RVE52072.1 KY564020 ASO76399.1 KM229767 AKC05613.1 GL451103 EFN79997.1 KQ971341 EFA04511.1 EFN80029.1 FX985621 BBF97952.1 KQ435726 KOX78148.1 AJ783863 CAH04311.1 GL763324 EFZ19789.1 GL444277 EFN61067.1 GBYB01006060 JAG75827.1 JQ771366 AFD93397.1 GALX01004089 JAB64377.1 KZ288381 PBC26515.1 JR041657 AEY59487.1 GDAI01001408 JAI16195.1 JQ771358 AFD93389.1 JQ771367 AFD93398.1 MH365708 QBB01517.1 FN908680 CBM69268.1 KQ434786 KZC05146.1 KK107538 QOIP01000005 EZA49150.1 RLU23180.1 KX447597 AOA60264.1 AJ783862 CAH04310.1 LBMM01005253 KMQ91679.1 GL888613 EGI58991.1 KQ976439 KYM86708.1 KQ981693 KYN37872.1 ADTU01018960 KQ982409 KYQ56817.1 DS232047 EDS33081.1 GFDL01009055 JAV25990.1 KQ977649 KYN00740.1 APGK01051867 BT126745 KB741207 KB632327 AEE61707.1 ENN72907.1 ERL92523.1 DQ440195 CH477695 ABF18228.1 EAT37166.1 KQ980886 KYN11839.1 BT127154 AEE62116.1 JXUM01082821 GAPW01002729 KQ563338 JAC10869.1 KXJ74024.1 AXCN02000015 GDHC01008765 JAQ09864.1 AXCM01001019 GGFM01000110 MBW20861.1 GGFM01000150 MBW20901.1 GGFJ01007529 MBW56670.1 GEZM01053565 JAV74013.1 GGFK01000447 MBW33768.1 ADMH02001019 ETN64428.1 GBXI01010522 JAD03770.1 AY127558 AAM97779.1 AJVK01011094 CH902618 EDV41295.1 APCN01005540 AAAB01008888 EAA08855.2 UFQS01000821 UFQT01000410 UFQT01000821 SSX07034.1 SSX24038.1 GAKP01019889 JAC39063.1 KK852865 KDR14762.1 CH916366 EDV97495.1 KC507365 AHB12488.1 GAMC01000654 JAC05902.1 Y18444 AF220939 AAF31449.1 CH379070 EAL30389.1 OUUW01000002 SPP76390.1 GDHF01029382 GDHF01028350 JAI22932.1 JAI23964.1 AE014296 AGB94895.1 M25772 AY075528 BT021447 CH954178 EDV52744.1 CM000363 EDX11475.1 CH480826 EDW44450.1 GANO01001021 JAB58850.1 CM002912 KMZ01149.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000037510
+ More
UP000283053 UP000008237 UP000007266 UP000053105 UP000002358 UP000000311 UP000242457 UP000005203 UP000076502 UP000053097 UP000279307 UP000036403 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000002320 UP000078542 UP000019118 UP000030742 UP000008820 UP000078492 UP000069940 UP000249989 UP000075886 UP000075883 UP000075881 UP000075900 UP000069272 UP000075880 UP000000673 UP000075920 UP000092462 UP000007801 UP000075882 UP000075902 UP000075840 UP000075903 UP000076407 UP000007062 UP000027135 UP000001070 UP000001819 UP000075884 UP000268350 UP000192221 UP000000803 UP000008711 UP000078200 UP000000304 UP000001292
UP000283053 UP000008237 UP000007266 UP000053105 UP000002358 UP000000311 UP000242457 UP000005203 UP000076502 UP000053097 UP000279307 UP000036403 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000002320 UP000078542 UP000019118 UP000030742 UP000008820 UP000078492 UP000069940 UP000249989 UP000075886 UP000075883 UP000075881 UP000075900 UP000069272 UP000075880 UP000000673 UP000075920 UP000092462 UP000007801 UP000075882 UP000075902 UP000075840 UP000075903 UP000076407 UP000007062 UP000027135 UP000001070 UP000001819 UP000075884 UP000268350 UP000192221 UP000000803 UP000008711 UP000078200 UP000000304 UP000001292
ProteinModelPortal
Q70YZ3
Q5UAU1
A0A212FEQ6
G0ZEC4
E7DZ05
I4DID8
+ More
A0A194RK76 A0A2A4J9D4 A0A0K8SL83 A0A0L7LDY8 D1LYK0 I4DLY9 A0A3G1T1B4 Q8WQJ2 A0A3S2LR49 A0A222AJ53 A0A185RX45 E2BW93 D6WMT4 E2BWC5 A0A348G679 A0A0N0BIW7 Q6EV08 K7IQV7 E9II14 E2AZT8 A0A0C9RCN3 I6P504 V5GRW6 A0A2A3E478 V9IFN9 A0A087ZPA5 A0A0K8TQM2 I6P155 I6P6U6 A0A411G6P8 E6ZCJ9 A0A154NZP4 A0A026VZT8 A0A1B2M4Y0 Q6EV09 A0A0J7KN60 F4X3G3 A0A195BNB3 A0A195FCE2 A0A158NKQ4 A0A151X8W5 B0WQZ4 A0A1Q3FEK8 A0A195CJ37 J3JU33 Q1HR99 A0A195DHA1 J3JV87 A0A023EQJ9 A0A182Q7D3 A0A146LPN5 A0A182LZB4 A0A182K8J7 A0A182RZI1 A0A2M3YX70 A0A182FNS5 A0A2M3YXC4 A0A2M4BUA9 A0A182IRR4 A0A1Y1LK51 A0A2M3ZZC9 W5JMB2 A0A1Y9IV09 A0A0A1WZ39 Q8MQT0 A0A1B0D3Y1 B3MAK8 A0A182KXJ9 A0A182U5N4 A0A182HZC3 A0A182V7K1 A0A182WVR6 Q7PQG7 A0A336MDC9 A0A034VBX9 A0A067R8K2 B4IXN3 A0A109NIB3 W8CB96 Q9U3U0 Q9NHP0 Q29DM5 A0A182N4R2 A0A3B0JLU0 A0A0K8UBE5 A0A1W4VAZ1 M9PG76 P19889 B3NJ27 A0A1A9UP79 B4QKR0 B4IAY7 U5EVH7 A0A0J9RZV4
A0A194RK76 A0A2A4J9D4 A0A0K8SL83 A0A0L7LDY8 D1LYK0 I4DLY9 A0A3G1T1B4 Q8WQJ2 A0A3S2LR49 A0A222AJ53 A0A185RX45 E2BW93 D6WMT4 E2BWC5 A0A348G679 A0A0N0BIW7 Q6EV08 K7IQV7 E9II14 E2AZT8 A0A0C9RCN3 I6P504 V5GRW6 A0A2A3E478 V9IFN9 A0A087ZPA5 A0A0K8TQM2 I6P155 I6P6U6 A0A411G6P8 E6ZCJ9 A0A154NZP4 A0A026VZT8 A0A1B2M4Y0 Q6EV09 A0A0J7KN60 F4X3G3 A0A195BNB3 A0A195FCE2 A0A158NKQ4 A0A151X8W5 B0WQZ4 A0A1Q3FEK8 A0A195CJ37 J3JU33 Q1HR99 A0A195DHA1 J3JV87 A0A023EQJ9 A0A182Q7D3 A0A146LPN5 A0A182LZB4 A0A182K8J7 A0A182RZI1 A0A2M3YX70 A0A182FNS5 A0A2M3YXC4 A0A2M4BUA9 A0A182IRR4 A0A1Y1LK51 A0A2M3ZZC9 W5JMB2 A0A1Y9IV09 A0A0A1WZ39 Q8MQT0 A0A1B0D3Y1 B3MAK8 A0A182KXJ9 A0A182U5N4 A0A182HZC3 A0A182V7K1 A0A182WVR6 Q7PQG7 A0A336MDC9 A0A034VBX9 A0A067R8K2 B4IXN3 A0A109NIB3 W8CB96 Q9U3U0 Q9NHP0 Q29DM5 A0A182N4R2 A0A3B0JLU0 A0A0K8UBE5 A0A1W4VAZ1 M9PG76 P19889 B3NJ27 A0A1A9UP79 B4QKR0 B4IAY7 U5EVH7 A0A0J9RZV4
PDB
5AJ0
E-value=6.56176e-106,
Score=980
Ontologies
GO
GO:0005840
GO:0042254
GO:0022625
GO:0002181
GO:0070180
GO:0003735
GO:0000027
GO:0006284
GO:0005509
GO:0052720
GO:0016363
GO:0005829
GO:0005654
GO:0000287
GO:0090305
GO:0003906
GO:0004519
GO:0140078
GO:0022626
GO:0005622
GO:0008168
GO:0005515
GO:0003676
GO:0000166
GO:0046872
GO:0016020
GO:0016876
GO:0043039
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
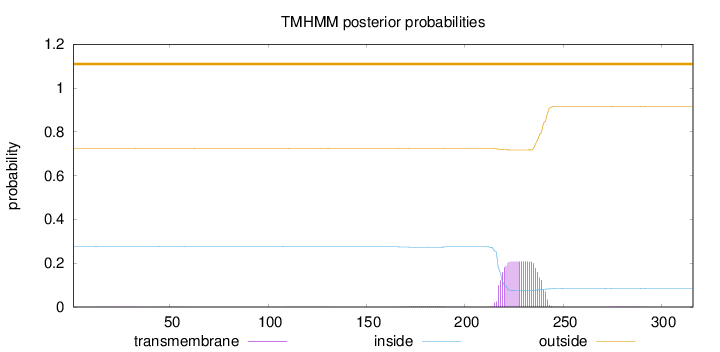
Length:
316
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.6004
Exp number, first 60 AAs:
0.00785
Total prob of N-in:
0.27582
outside
1 - 316
Population Genetic Test Statistics
Pi
14.404165
Theta
15.870635
Tajima's D
-0.593822
CLR
0.169589
CSRT
0.219589020548973
Interpretation
Uncertain
