Gene
KWMTBOMO09298
Pre Gene Modal
BGIBMGA003433
Annotation
RCC1_domain-containing_protein_1_[Operophtera_brumata]
Location in the cell
Extracellular Reliability : 1.168 Nuclear Reliability : 1.105
Sequence
CDS
ATGTCTAAATCTGAAGATCCAAATTTTGACTTTAGATTGGCTAATTTAATGCAAATCAATTGGTCATATAATATATTTCAACTAAGAAAAAATCTTTATTTATTGGGCGCCTTTGACGGTAGGAAAATCGCTAAGAAAATTGATATACCAGAAGAGTGCAGAAAATCGCCACTCGTTGCCGGTAACGACTACAACTTGGTTATTGGCCACAATAATTCGAGTGCCCTTTGGATATTGAATTTGGAAGATAATAGCATAAAGAATGTTTATTTGGATAGAGAATTGCCTCTTGAATTATCGCCCAAACGTAATAAAACAGATAATGAGATACAAAAGCTTGTTGTTAATAATTTTTCATGCCTCTACCTTACTAAAGGCAAGAATGTTTATAGCGGAATACTTCCAAGTTACATAGATACAAGCCACTGTAATGGACATATTATTGATGTTGCTTGTGGTTATGAACATTACATGCTTCTCACTAGTACTGGTTATGTCTATACTTGGGGAGATGGCAGGAGATTGCAACTAGGTCAGGATGATACAAGCAATTTAGAGACACCCACACAGGTTGAAGCCCTGGCTGGAATAAAAATTATTAAAATAAAAGCAGGTGGGTGGCATTCCATGGCACTCAGTGAATTTGGGGATCTCTATGTTTGGGGCTTGAATGACACTGGTCAATTAGGTATAAAGGAAAGTGAAAATGACTTAAAAAGCTATGCTTTACCAAAGTTAGTTGATCTATATGATGATAATGAAGAAGTTCATCTCCAAATTAAGGACATAGCCTGTGGCTCGAAGCATTCTGTATTATTATTAGAAGATGGCTCAGTTTGGACCACTGGTGCAAATAAATATGGACAACTAGCATGCCCACAATATCCAAACTCAAAAAACTTCAATAAGGTCTGTGTAATCAAAGAAGCATGTAGACTGAAATGTGGACCATGGACAACTGTAGTTGTATGCAATTAA
Protein
MSKSEDPNFDFRLANLMQINWSYNIFQLRKNLYLLGAFDGRKIAKKIDIPEECRKSPLVAGNDYNLVIGHNNSSALWILNLEDNSIKNVYLDRELPLELSPKRNKTDNEIQKLVVNNFSCLYLTKGKNVYSGILPSYIDTSHCNGHIIDVACGYEHYMLLTSTGYVYTWGDGRRLQLGQDDTSNLETPTQVEALAGIKIIKIKAGGWHSMALSEFGDLYVWGLNDTGQLGIKESENDLKSYALPKLVDLYDDNEEVHLQIKDIACGSKHSVLLLEDGSVWTTGANKYGQLACPQYPNSKNFNKVCVIKEACRLKCGPWTTVVVCN
Summary
Uniprot
H9J1P6
A0A2H1V8T6
A0A0L7LD68
A0A2A4J9K9
A0A1E1WCX6
A0A3S2TQ45
+ More
S4PRX1 A0A194PYI2 A0A194QUN2 A0A1B6JUZ2 A0A1B6F374 A0A154PSS9 A0A1B6LQS4 A0A0J7KB09 E2AS43 A0A195E3W8 A0A067RE98 A0A195BSB1 A0A1I8Q0J4 A0A158NXY1 A0A0L7QKR4 E2B430 A0A151WQY4 A0A2A3EV34 F4X4D0 A0A088AFD2 A0A0N8APE8 B4KAP1 A0A164QJE0 A0A151IGS5 K7JJB0 A0A0K8UPF0 B3LVJ5 A0A1W4VRG3 A0A1A9W5P2 A0A1I8MYC4 A0A026WA86 E9ICC6 A0A034V9W0 A0A2J7RR47 A0A2J7RR60 A0A2J7RR49 A0A2J7RR56 A0A0A1WQ12 A0A0M3QYC7 A0A1A9ZSV5 T1KZX9 A0A1A9VDQ0 W8AUZ1 A0A1A9Y0I9 A0A1W4W453 B4JRK5 A0A1B0ETG2 B4NHQ3 A0A0Q9WR27 B4M0P2 A0A1B0B2Y6 A0A0Q9WGB9 B3P8Q4 B4G644 A0A3B0JL36 Q29AP8 A0A443SQF2 B4QZB6 A0A132AB82
S4PRX1 A0A194PYI2 A0A194QUN2 A0A1B6JUZ2 A0A1B6F374 A0A154PSS9 A0A1B6LQS4 A0A0J7KB09 E2AS43 A0A195E3W8 A0A067RE98 A0A195BSB1 A0A1I8Q0J4 A0A158NXY1 A0A0L7QKR4 E2B430 A0A151WQY4 A0A2A3EV34 F4X4D0 A0A088AFD2 A0A0N8APE8 B4KAP1 A0A164QJE0 A0A151IGS5 K7JJB0 A0A0K8UPF0 B3LVJ5 A0A1W4VRG3 A0A1A9W5P2 A0A1I8MYC4 A0A026WA86 E9ICC6 A0A034V9W0 A0A2J7RR47 A0A2J7RR60 A0A2J7RR49 A0A2J7RR56 A0A0A1WQ12 A0A0M3QYC7 A0A1A9ZSV5 T1KZX9 A0A1A9VDQ0 W8AUZ1 A0A1A9Y0I9 A0A1W4W453 B4JRK5 A0A1B0ETG2 B4NHQ3 A0A0Q9WR27 B4M0P2 A0A1B0B2Y6 A0A0Q9WGB9 B3P8Q4 B4G644 A0A3B0JL36 Q29AP8 A0A443SQF2 B4QZB6 A0A132AB82
Pubmed
EMBL
BABH01023977
ODYU01001255
SOQ37211.1
JTDY01001598
KOB73417.1
NWSH01002527
+ More
PCG68083.1 GDQN01006249 JAT84805.1 RSAL01000026 RVE52070.1 GAIX01000440 JAA92120.1 KQ459594 KPI96210.1 KQ461108 KPJ09177.1 GECU01022006 GECU01004681 JAS85700.1 JAT03026.1 GECZ01025109 JAS44660.1 KQ435180 KZC14969.1 GEBQ01014038 JAT25939.1 LBMM01010461 KMQ87446.1 GL442221 EFN63743.1 KQ979685 KYN19776.1 KK852521 KDR22087.1 KQ976423 KYM88892.1 ADTU01000542 KQ414940 KOC59233.1 GL445463 EFN89544.1 KQ982815 KYQ50218.1 KZ288185 PBC34931.1 GL888633 EGI58726.1 GDIQ01255875 JAJ95849.1 CH933806 EDW16778.1 LRGB01002384 KZS07808.1 KQ977671 KYN00621.1 GDHF01023853 JAI28461.1 CH902617 EDV42565.1 KK107323 EZA52561.1 GL762231 EFZ21794.1 GAKP01019665 GAKP01019663 JAC39289.1 NEVH01000611 PNF43310.1 PNF43313.1 PNF43316.1 PNF43314.1 GBXI01013365 JAD00927.1 CP012526 ALC47389.1 CAEY01000758 GAMC01013850 GAMC01013848 JAB92705.1 CH916373 EDV94395.1 CCAG010010758 CH964272 EDW84663.2 CH940650 KRF83252.1 EDW67334.2 JXJN01007739 KRF83250.1 KRF83251.1 CH954182 EDV54218.1 CH479179 EDW23797.1 OUUW01000007 SPP82935.1 CM000070 EAL27301.2 NCKV01000786 RWS29744.1 CM000364 EDX13852.1 JXLN01012231 KPM08167.1
PCG68083.1 GDQN01006249 JAT84805.1 RSAL01000026 RVE52070.1 GAIX01000440 JAA92120.1 KQ459594 KPI96210.1 KQ461108 KPJ09177.1 GECU01022006 GECU01004681 JAS85700.1 JAT03026.1 GECZ01025109 JAS44660.1 KQ435180 KZC14969.1 GEBQ01014038 JAT25939.1 LBMM01010461 KMQ87446.1 GL442221 EFN63743.1 KQ979685 KYN19776.1 KK852521 KDR22087.1 KQ976423 KYM88892.1 ADTU01000542 KQ414940 KOC59233.1 GL445463 EFN89544.1 KQ982815 KYQ50218.1 KZ288185 PBC34931.1 GL888633 EGI58726.1 GDIQ01255875 JAJ95849.1 CH933806 EDW16778.1 LRGB01002384 KZS07808.1 KQ977671 KYN00621.1 GDHF01023853 JAI28461.1 CH902617 EDV42565.1 KK107323 EZA52561.1 GL762231 EFZ21794.1 GAKP01019665 GAKP01019663 JAC39289.1 NEVH01000611 PNF43310.1 PNF43313.1 PNF43316.1 PNF43314.1 GBXI01013365 JAD00927.1 CP012526 ALC47389.1 CAEY01000758 GAMC01013850 GAMC01013848 JAB92705.1 CH916373 EDV94395.1 CCAG010010758 CH964272 EDW84663.2 CH940650 KRF83252.1 EDW67334.2 JXJN01007739 KRF83250.1 KRF83251.1 CH954182 EDV54218.1 CH479179 EDW23797.1 OUUW01000007 SPP82935.1 CM000070 EAL27301.2 NCKV01000786 RWS29744.1 CM000364 EDX13852.1 JXLN01012231 KPM08167.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000076502 UP000036403 UP000000311 UP000078492 UP000027135 UP000078540 UP000095300 UP000005205 UP000053825 UP000008237 UP000075809 UP000242457 UP000007755 UP000005203 UP000009192 UP000076858 UP000078542 UP000002358 UP000007801 UP000192221 UP000091820 UP000095301 UP000053097 UP000235965 UP000092553 UP000092445 UP000015104 UP000078200 UP000092443 UP000001070 UP000092444 UP000007798 UP000008792 UP000092460 UP000008711 UP000008744 UP000268350 UP000001819 UP000288716 UP000000304
UP000076502 UP000036403 UP000000311 UP000078492 UP000027135 UP000078540 UP000095300 UP000005205 UP000053825 UP000008237 UP000075809 UP000242457 UP000007755 UP000005203 UP000009192 UP000076858 UP000078542 UP000002358 UP000007801 UP000192221 UP000091820 UP000095301 UP000053097 UP000235965 UP000092553 UP000092445 UP000015104 UP000078200 UP000092443 UP000001070 UP000092444 UP000007798 UP000008792 UP000092460 UP000008711 UP000008744 UP000268350 UP000001819 UP000288716 UP000000304
PRIDE
Pfam
PF00415 RCC1
SUPFAM
SSF50985
SSF50985
Gene 3D
ProteinModelPortal
H9J1P6
A0A2H1V8T6
A0A0L7LD68
A0A2A4J9K9
A0A1E1WCX6
A0A3S2TQ45
+ More
S4PRX1 A0A194PYI2 A0A194QUN2 A0A1B6JUZ2 A0A1B6F374 A0A154PSS9 A0A1B6LQS4 A0A0J7KB09 E2AS43 A0A195E3W8 A0A067RE98 A0A195BSB1 A0A1I8Q0J4 A0A158NXY1 A0A0L7QKR4 E2B430 A0A151WQY4 A0A2A3EV34 F4X4D0 A0A088AFD2 A0A0N8APE8 B4KAP1 A0A164QJE0 A0A151IGS5 K7JJB0 A0A0K8UPF0 B3LVJ5 A0A1W4VRG3 A0A1A9W5P2 A0A1I8MYC4 A0A026WA86 E9ICC6 A0A034V9W0 A0A2J7RR47 A0A2J7RR60 A0A2J7RR49 A0A2J7RR56 A0A0A1WQ12 A0A0M3QYC7 A0A1A9ZSV5 T1KZX9 A0A1A9VDQ0 W8AUZ1 A0A1A9Y0I9 A0A1W4W453 B4JRK5 A0A1B0ETG2 B4NHQ3 A0A0Q9WR27 B4M0P2 A0A1B0B2Y6 A0A0Q9WGB9 B3P8Q4 B4G644 A0A3B0JL36 Q29AP8 A0A443SQF2 B4QZB6 A0A132AB82
S4PRX1 A0A194PYI2 A0A194QUN2 A0A1B6JUZ2 A0A1B6F374 A0A154PSS9 A0A1B6LQS4 A0A0J7KB09 E2AS43 A0A195E3W8 A0A067RE98 A0A195BSB1 A0A1I8Q0J4 A0A158NXY1 A0A0L7QKR4 E2B430 A0A151WQY4 A0A2A3EV34 F4X4D0 A0A088AFD2 A0A0N8APE8 B4KAP1 A0A164QJE0 A0A151IGS5 K7JJB0 A0A0K8UPF0 B3LVJ5 A0A1W4VRG3 A0A1A9W5P2 A0A1I8MYC4 A0A026WA86 E9ICC6 A0A034V9W0 A0A2J7RR47 A0A2J7RR60 A0A2J7RR49 A0A2J7RR56 A0A0A1WQ12 A0A0M3QYC7 A0A1A9ZSV5 T1KZX9 A0A1A9VDQ0 W8AUZ1 A0A1A9Y0I9 A0A1W4W453 B4JRK5 A0A1B0ETG2 B4NHQ3 A0A0Q9WR27 B4M0P2 A0A1B0B2Y6 A0A0Q9WGB9 B3P8Q4 B4G644 A0A3B0JL36 Q29AP8 A0A443SQF2 B4QZB6 A0A132AB82
PDB
3KCI
E-value=1.14992e-14,
Score=193
Ontologies
GO
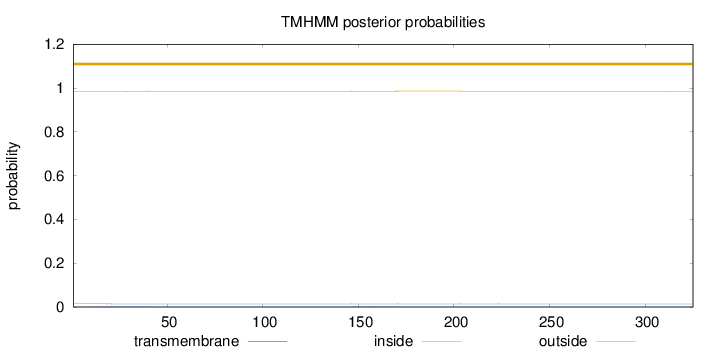
Topology
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0373399999999999
Exp number, first 60 AAs:
0.02083
Total prob of N-in:
0.01501
outside
1 - 325
Population Genetic Test Statistics
Pi
249.821121
Theta
210.674193
Tajima's D
0.585008
CLR
0
CSRT
0.534823258837058
Interpretation
Uncertain
