Pre Gene Modal
BGIBMGA003428
Annotation
PREDICTED:_dolichyl-diphosphooligosaccharide--protein_glycosyltransferase_subunit_STT3A_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.949
Sequence
CDS
ATGACGGCGGAAACACAGAGCAAGTCCAGATTACCGTATTTGTCTTCGGACAAACAACTAACATTTATAAAACTCTCTGTATTGTCAATGGCAGCAATTTTATCTTTTGCGACGCGTTTATTCTCCGTTCTGCGTTTTGAGAGCGTAATCCATGAGTTTGATCCCTACTTCAACTATAGAACGACGAGATATCTCACTGAAGAGGGATTTTATAACTTCCACAACTGGTTTGACGATAGAGCATGGTATCCTCTAGGTCGTATTATTGGAGGTACCATCTACCCTGGCCTGATGGTGACTTCTGCCACTCTTTACAATATAATGCAGTACTTAAACATAACAATTGACATCAGAAATGTATGTGTGTTCTTGGCTCCATTCTTCTCCTCACTCACAACTATTGTCACATACCTCCTTACCAAGGAGTTAAAGAATGAAGGAGCGGGCTTAGTGGCAGCAGCAATGATTGCAATCGTCCCTGGTTACATCAGTCGTTCAGTCGCAGGAAGCTATGATAATGAAGGCATCGCCATCTTCTGCATGCTGCTGACTTATTATTTTTGGATCAAAGCTGTTAATACAGGAACTATCATGTGGGCAACTATGACGGCTCTGGCATATTTCTACATGGTGTCATCATGGGGTGGCTACGTTTTCCTCATAAACCTGATACCGTTGCACGTACTGGCCCTCATTCTGCTGGGAAGGTTCTCGTCGAGGGTGTACGTGGCTTACAGCACGCTGTACTGCATCGGAACCATCCTCTCAATGCAGATTTCTTTTGTGGGCTTCCAACCGGTGCAGAGTTCGGAGCATATGTTGGCTCTCGGTACATTCGGGCTGTGTCAGCTGTACGCGTTCACGCAGTACCTCAGACAACATCTGTCGCCGGCTAACTTCGAGCTGCTCTTCAAGGCTCTTGTTACAACGCTGCTTCTGACCCTGGGCTCTGCTATCATAGCCCTCACTTTGACCGGAAAAATCTCACCGTGGACTGGCCGTTTCTACTCATTGCTCGATCCATCGTACGCTAAGAACCACATTCCTATCATTGCATCCGTGTCCGAGCATCAGCCCACGTCGTGGTCATCATTCTACTTCGACCTCCAAGTCCTGGTGTTCCTGTTCCCGGCTGGACTGTACTTTTGCTTTAGCAAGCTCACGGATGCAAACATATTCATCATTCTGTATGGTGTGCTCAGTATATACTTTGCTGGCGTCATGGTTCGTCTTATGCTGGTACTCGCTCCGGTCATGTGCATCGTCTCCGGTATAGCAGCTTCCAGTCTACTTAGTCTTCACGTTAAGGATATTGAGCCTAAAGTTGAGAAACACGATAAGAAGAAGAAGCATGAAAATAATCTTGTGTTTAGATCTGAGGTGGGCACGTTGTTCGTGTGCGTGCTGGGCTGCCTGCTGGTGTCGTACGTGGTGCACTGCACGTGGGTCACGTCGGAGGCCTACTCGTCGCCCTCCATCGTGCTGTCGGCGCGCGCACACGACGGCGCCCGGATCATCTTCGACGACTTCCGCGAGGCCTACACCTGGCTCAAGATGAACACTCCAGAGGACGCCAAAGTGATGTCGTGGTGGGACTACGGCTACCAAATAACGGCGATGGCGAACCGGACGGTGATCGTCGACAACAACACGTGGAACAACACGCACATATCGCGCGTGGGGCAGGCGATGGCCTCCACCGAGGACAAGGCCTACGAGATCATGAGGGAGCTGGACGTGGACTACGTGCTCGTCATCTTCGGAGGACTGGTCGGATACTCTTCGGACGACATCAACAAGTTCCTGTGGATGGTGCGCATCGGCGGCAGCACGGAGCGCGGCGCGCACATCCGCGAGGCGCAGTACTACACGGCGGCCGGGGAGTTCCGCGTGGACTCGGCCGGCGCGCCCGCGCTGCTCAACTGCCTCATGTACAAGATGAGCTACTACAAGTTCGGACTCGTCTACACGGAGGGCGGCCGGCCGCCAGGGTTCGATCGGGTTCGAGGAGCTGAGATCGGCAACAAGGACTTCAATCTAGATGTTCTAGAGGAAGCCTACACGACAGAACATTGGCTGGTCCGCATTTACAAAGTCAAACCTTTGCCGAACAGGGGAGTATAA
Protein
MTAETQSKSRLPYLSSDKQLTFIKLSVLSMAAILSFATRLFSVLRFESVIHEFDPYFNYRTTRYLTEEGFYNFHNWFDDRAWYPLGRIIGGTIYPGLMVTSATLYNIMQYLNITIDIRNVCVFLAPFFSSLTTIVTYLLTKELKNEGAGLVAAAMIAIVPGYISRSVAGSYDNEGIAIFCMLLTYYFWIKAVNTGTIMWATMTALAYFYMVSSWGGYVFLINLIPLHVLALILLGRFSSRVYVAYSTLYCIGTILSMQISFVGFQPVQSSEHMLALGTFGLCQLYAFTQYLRQHLSPANFELLFKALVTTLLLTLGSAIIALTLTGKISPWTGRFYSLLDPSYAKNHIPIIASVSEHQPTSWSSFYFDLQVLVFLFPAGLYFCFSKLTDANIFIILYGVLSIYFAGVMVRLMLVLAPVMCIVSGIAASSLLSLHVKDIEPKVEKHDKKKKHENNLVFRSEVGTLFVCVLGCLLVSYVVHCTWVTSEAYSSPSIVLSARAHDGARIIFDDFREAYTWLKMNTPEDAKVMSWWDYGYQITAMANRTVIVDNNTWNNTHISRVGQAMASTEDKAYEIMRELDVDYVLVIFGGLVGYSSDDINKFLWMVRIGGSTERGAHIREAQYYTAAGEFRVDSAGAPALLNCLMYKMSYYKFGLVYTEGGRPPGFDRVRGAEIGNKDFNLDVLEEAYTTEHWLVRIYKVKPLPNRGV
Summary
Uniprot
H9J1P1
A0A194RJU0
A0A2W1BVJ8
A0A194PKS6
A0A3S2M5U2
A0A2A4J9P5
+ More
A0A2H1VSY6 A0A212EGW9 A0A1B6HC36 S4NTV6 A0A067RGL4 A0A2J7PE11 A0A224XKS3 A0A0N7Z980 A0A069DWJ8 R4FP94 A0A0A9YZB0 A0A023F931 A0A1Y1LEM1 A0A1B6DKE0 A0A0K8TMR8 Q17DE3 A0A1Q3F4A6 A0A0A1WZY7 A0A0K8UYU3 A0A034VB92 A0A182HAS4 A0A1A9UEI2 A0A1B0ADN2 A0A0P6CDI1 A0A1B0FQ19 A0A0L0BMJ6 E9G6L8 A0A0P5FVY6 A0A3R7QMB5 W5JFC1 A0A182IY80 A0A0N8AY16 A0A061QLL8 A0A0P5Q7K3 A0A0P5NU05 A0A182W5G6 A0A1A9XSL1 A0A1B0AQG1 B4PYM0 B3NY63 W8BAI9 B3DN57 B4L8Q4 Q9VRE0 A0A084VLE4 U5EW59 A0A0N8DUS9 A0A1A9W8N8 A0A182RMW2 D6WKL9 A0A0N8EE82 A0A0P5QHP2 A0A026W473 A0A1W4VWQ0 A0A182XXU9 A0A1I8Q0G6 A0A0P6EJG1 B4NCR5 A0A182SYJ8 B4M1T6 A0A1I8MLD4 A0A182UEW2 Q7PS25 A0A182VGX2 B4I6D5 A0A182PBK1 A0A3L8E1R2 B3N0J0 A0A3B0J887 A0A0N0U4L1 A0A0P5IFR8 A0A1W4WSH1 B4JIP5 A0A182GQJ9 A0A087ZWD2 A0A310SNA2 F4W854 A0A195EYT6 A0A158NDQ5 A0A151X418 A0A0M5J0V3 A0A154PSC3 A0A0L7QV25 E2A0N1 A0A2M4A226 A0A034VC64 B5DNT0 A0A2R5LE35 A0A2A3ELE9 A0A293N280 A0A232FJU7 K7JBD1 A0A1B6L1G3 A0A0C9RC03
A0A2H1VSY6 A0A212EGW9 A0A1B6HC36 S4NTV6 A0A067RGL4 A0A2J7PE11 A0A224XKS3 A0A0N7Z980 A0A069DWJ8 R4FP94 A0A0A9YZB0 A0A023F931 A0A1Y1LEM1 A0A1B6DKE0 A0A0K8TMR8 Q17DE3 A0A1Q3F4A6 A0A0A1WZY7 A0A0K8UYU3 A0A034VB92 A0A182HAS4 A0A1A9UEI2 A0A1B0ADN2 A0A0P6CDI1 A0A1B0FQ19 A0A0L0BMJ6 E9G6L8 A0A0P5FVY6 A0A3R7QMB5 W5JFC1 A0A182IY80 A0A0N8AY16 A0A061QLL8 A0A0P5Q7K3 A0A0P5NU05 A0A182W5G6 A0A1A9XSL1 A0A1B0AQG1 B4PYM0 B3NY63 W8BAI9 B3DN57 B4L8Q4 Q9VRE0 A0A084VLE4 U5EW59 A0A0N8DUS9 A0A1A9W8N8 A0A182RMW2 D6WKL9 A0A0N8EE82 A0A0P5QHP2 A0A026W473 A0A1W4VWQ0 A0A182XXU9 A0A1I8Q0G6 A0A0P6EJG1 B4NCR5 A0A182SYJ8 B4M1T6 A0A1I8MLD4 A0A182UEW2 Q7PS25 A0A182VGX2 B4I6D5 A0A182PBK1 A0A3L8E1R2 B3N0J0 A0A3B0J887 A0A0N0U4L1 A0A0P5IFR8 A0A1W4WSH1 B4JIP5 A0A182GQJ9 A0A087ZWD2 A0A310SNA2 F4W854 A0A195EYT6 A0A158NDQ5 A0A151X418 A0A0M5J0V3 A0A154PSC3 A0A0L7QV25 E2A0N1 A0A2M4A226 A0A034VC64 B5DNT0 A0A2R5LE35 A0A2A3ELE9 A0A293N280 A0A232FJU7 K7JBD1 A0A1B6L1G3 A0A0C9RC03
Pubmed
19121390
26354079
28756777
22118469
23622113
24845553
+ More
27129103 26334808 25401762 26823975 25474469 28004739 26369729 17510324 25830018 25348373 26483478 26108605 21292972 20920257 23761445 17994087 17550304 24495485 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 18362917 19820115 24508170 25244985 25315136 12364791 14747013 17210077 30249741 21719571 21347285 20798317 15632085 23185243 28648823 20075255
27129103 26334808 25401762 26823975 25474469 28004739 26369729 17510324 25830018 25348373 26483478 26108605 21292972 20920257 23761445 17994087 17550304 24495485 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 18362917 19820115 24508170 25244985 25315136 12364791 14747013 17210077 30249741 21719571 21347285 20798317 15632085 23185243 28648823 20075255
EMBL
BABH01023963
KQ460106
KPJ17817.1
KZ149923
PZC77654.1
KQ459602
+ More
KPI93603.1 RSAL01000026 RVE52063.1 NWSH01002254 PCG68805.1 ODYU01004261 SOQ43943.1 AGBW02015002 OWR40733.1 GECU01035466 JAS72240.1 GAIX01012071 JAA80489.1 KK852696 KDR18262.1 NEVH01026112 PNF14573.1 GFTR01007795 JAW08631.1 GDKW01001291 JAI55304.1 GBGD01000673 JAC88216.1 ACPB03005610 GAHY01000994 JAA76516.1 GBHO01006080 GDHC01021415 JAG37524.1 JAP97213.1 GBBI01000731 JAC17981.1 GEZM01062156 GEZM01062155 JAV70006.1 GEDC01011139 JAS26159.1 GDAI01001956 JAI15647.1 CH477296 EAT44394.1 GFDL01012654 JAV22391.1 GBXI01010334 JAD03958.1 GDHF01026345 GDHF01021502 GDHF01020799 JAI25969.1 JAI30812.1 JAI31515.1 GAKP01019581 GAKP01019580 GAKP01019579 JAC39372.1 JXUM01031403 JXUM01031404 JXUM01031405 KQ560920 KXJ80345.1 GDIP01009873 LRGB01000512 JAM93842.1 KZS18894.1 CCAG010020168 JRES01001642 KNC21261.1 GL732533 EFX84969.1 GDIQ01253686 GDIQ01253685 GDIQ01250989 GDIQ01247842 GDIQ01247841 GDIQ01243801 GDIQ01190607 GDIQ01190606 GDIQ01187906 GDIQ01186358 GDIQ01184710 GDIQ01178204 GDIQ01169506 GDIQ01166911 GDIQ01155622 GDIQ01148333 GDIQ01146872 GDIQ01140885 GDIQ01132492 GDIQ01132491 GDIQ01129888 GDIQ01127245 GDIQ01125532 GDIQ01123678 GDIQ01119271 GDIQ01119270 GDIQ01113587 GDIQ01100842 GDIQ01099431 GDIQ01099430 GDIQ01087842 GDIQ01087841 GDIQ01084602 GDIQ01079936 GDIQ01077987 GDIQ01050966 GDIQ01050965 GDIQ01049714 GDIQ01048211 GDIQ01046460 GDIQ01044469 JAK00736.1 QCYY01000659 ROT83643.1 ADMH02001666 ETN61504.1 GDIQ01231731 JAK19994.1 GBFC01000029 JAC59309.1 GDIQ01121085 GDIQ01121084 JAL30641.1 GDIQ01137774 JAL13952.1 JXJN01001849 JXJN01001850 CM000162 EDX03061.1 CH954180 EDV47584.1 GAMC01016454 GAMC01016453 JAB90101.1 BT032845 ACD81859.1 CH933815 EDW08029.1 KRG07558.1 KRG07559.1 AE014298 AAF50861.1 AGB95589.2 ATLV01014483 KE524974 KFB38788.1 GANO01001569 JAB58302.1 GDIQ01089836 JAN04901.1 KQ971342 EFA03010.1 GDIQ01035412 JAN59325.1 GDIQ01135120 JAL16606.1 KK107436 EZA50852.1 GDIQ01075487 JAN19250.1 CH964239 EDW82624.1 CH940651 EDW65640.1 AAAB01008846 EAA06273.4 CH480823 EDW56341.1 QOIP01000002 RLU26129.1 CH902640 EDV38394.1 OUUW01000003 SPP78055.1 KQ435821 KOX72428.1 GDIQ01214001 JAK37724.1 CH916370 EDW00492.1 JXUM01080880 JXUM01080881 KQ563214 KXJ74280.1 KQ759851 OAD62552.1 GL887888 EGI69724.1 KQ981906 KYN33291.1 ADTU01012795 KQ982548 KYQ55163.1 CP012528 ALC48548.1 KQ435127 KZC14801.1 KQ414727 KOC62475.1 GL435626 EFN72934.1 GGFK01001484 MBW34805.1 GAKP01019582 JAC39370.1 CH379065 EDY73182.1 KRT07199.1 GGLE01003620 MBY07746.1 KZ288215 PBC32548.1 GFWV01022456 MAA47183.1 NNAY01000097 OXU31004.1 AAZX01003235 AAZX01010155 AAZX01012906 GEBQ01022482 JAT17495.1 GBYB01008133 GBYB01014114 JAG77900.1 JAG83881.1
KPI93603.1 RSAL01000026 RVE52063.1 NWSH01002254 PCG68805.1 ODYU01004261 SOQ43943.1 AGBW02015002 OWR40733.1 GECU01035466 JAS72240.1 GAIX01012071 JAA80489.1 KK852696 KDR18262.1 NEVH01026112 PNF14573.1 GFTR01007795 JAW08631.1 GDKW01001291 JAI55304.1 GBGD01000673 JAC88216.1 ACPB03005610 GAHY01000994 JAA76516.1 GBHO01006080 GDHC01021415 JAG37524.1 JAP97213.1 GBBI01000731 JAC17981.1 GEZM01062156 GEZM01062155 JAV70006.1 GEDC01011139 JAS26159.1 GDAI01001956 JAI15647.1 CH477296 EAT44394.1 GFDL01012654 JAV22391.1 GBXI01010334 JAD03958.1 GDHF01026345 GDHF01021502 GDHF01020799 JAI25969.1 JAI30812.1 JAI31515.1 GAKP01019581 GAKP01019580 GAKP01019579 JAC39372.1 JXUM01031403 JXUM01031404 JXUM01031405 KQ560920 KXJ80345.1 GDIP01009873 LRGB01000512 JAM93842.1 KZS18894.1 CCAG010020168 JRES01001642 KNC21261.1 GL732533 EFX84969.1 GDIQ01253686 GDIQ01253685 GDIQ01250989 GDIQ01247842 GDIQ01247841 GDIQ01243801 GDIQ01190607 GDIQ01190606 GDIQ01187906 GDIQ01186358 GDIQ01184710 GDIQ01178204 GDIQ01169506 GDIQ01166911 GDIQ01155622 GDIQ01148333 GDIQ01146872 GDIQ01140885 GDIQ01132492 GDIQ01132491 GDIQ01129888 GDIQ01127245 GDIQ01125532 GDIQ01123678 GDIQ01119271 GDIQ01119270 GDIQ01113587 GDIQ01100842 GDIQ01099431 GDIQ01099430 GDIQ01087842 GDIQ01087841 GDIQ01084602 GDIQ01079936 GDIQ01077987 GDIQ01050966 GDIQ01050965 GDIQ01049714 GDIQ01048211 GDIQ01046460 GDIQ01044469 JAK00736.1 QCYY01000659 ROT83643.1 ADMH02001666 ETN61504.1 GDIQ01231731 JAK19994.1 GBFC01000029 JAC59309.1 GDIQ01121085 GDIQ01121084 JAL30641.1 GDIQ01137774 JAL13952.1 JXJN01001849 JXJN01001850 CM000162 EDX03061.1 CH954180 EDV47584.1 GAMC01016454 GAMC01016453 JAB90101.1 BT032845 ACD81859.1 CH933815 EDW08029.1 KRG07558.1 KRG07559.1 AE014298 AAF50861.1 AGB95589.2 ATLV01014483 KE524974 KFB38788.1 GANO01001569 JAB58302.1 GDIQ01089836 JAN04901.1 KQ971342 EFA03010.1 GDIQ01035412 JAN59325.1 GDIQ01135120 JAL16606.1 KK107436 EZA50852.1 GDIQ01075487 JAN19250.1 CH964239 EDW82624.1 CH940651 EDW65640.1 AAAB01008846 EAA06273.4 CH480823 EDW56341.1 QOIP01000002 RLU26129.1 CH902640 EDV38394.1 OUUW01000003 SPP78055.1 KQ435821 KOX72428.1 GDIQ01214001 JAK37724.1 CH916370 EDW00492.1 JXUM01080880 JXUM01080881 KQ563214 KXJ74280.1 KQ759851 OAD62552.1 GL887888 EGI69724.1 KQ981906 KYN33291.1 ADTU01012795 KQ982548 KYQ55163.1 CP012528 ALC48548.1 KQ435127 KZC14801.1 KQ414727 KOC62475.1 GL435626 EFN72934.1 GGFK01001484 MBW34805.1 GAKP01019582 JAC39370.1 CH379065 EDY73182.1 KRT07199.1 GGLE01003620 MBY07746.1 KZ288215 PBC32548.1 GFWV01022456 MAA47183.1 NNAY01000097 OXU31004.1 AAZX01003235 AAZX01010155 AAZX01012906 GEBQ01022482 JAT17495.1 GBYB01008133 GBYB01014114 JAG77900.1 JAG83881.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000027135 UP000235965 UP000015103 UP000008820 UP000069940 UP000249989 UP000078200 UP000092445 UP000076858 UP000092444 UP000037069 UP000000305 UP000283509 UP000000673 UP000075880 UP000075920 UP000092443 UP000092460 UP000002282 UP000008711 UP000009192 UP000000803 UP000030765 UP000091820 UP000075900 UP000007266 UP000053097 UP000192221 UP000076408 UP000095300 UP000007798 UP000075901 UP000008792 UP000095301 UP000075902 UP000007062 UP000075903 UP000001292 UP000075885 UP000279307 UP000007801 UP000268350 UP000053105 UP000192223 UP000001070 UP000005203 UP000007755 UP000078541 UP000005205 UP000075809 UP000092553 UP000076502 UP000053825 UP000000311 UP000001819 UP000242457 UP000215335 UP000002358
UP000027135 UP000235965 UP000015103 UP000008820 UP000069940 UP000249989 UP000078200 UP000092445 UP000076858 UP000092444 UP000037069 UP000000305 UP000283509 UP000000673 UP000075880 UP000075920 UP000092443 UP000092460 UP000002282 UP000008711 UP000009192 UP000000803 UP000030765 UP000091820 UP000075900 UP000007266 UP000053097 UP000192221 UP000076408 UP000095300 UP000007798 UP000075901 UP000008792 UP000095301 UP000075902 UP000007062 UP000075903 UP000001292 UP000075885 UP000279307 UP000007801 UP000268350 UP000053105 UP000192223 UP000001070 UP000005203 UP000007755 UP000078541 UP000005205 UP000075809 UP000092553 UP000076502 UP000053825 UP000000311 UP000001819 UP000242457 UP000215335 UP000002358
Pfam
PF02516 STT3
Interpro
IPR003674
Oligo_trans_STT3
ProteinModelPortal
H9J1P1
A0A194RJU0
A0A2W1BVJ8
A0A194PKS6
A0A3S2M5U2
A0A2A4J9P5
+ More
A0A2H1VSY6 A0A212EGW9 A0A1B6HC36 S4NTV6 A0A067RGL4 A0A2J7PE11 A0A224XKS3 A0A0N7Z980 A0A069DWJ8 R4FP94 A0A0A9YZB0 A0A023F931 A0A1Y1LEM1 A0A1B6DKE0 A0A0K8TMR8 Q17DE3 A0A1Q3F4A6 A0A0A1WZY7 A0A0K8UYU3 A0A034VB92 A0A182HAS4 A0A1A9UEI2 A0A1B0ADN2 A0A0P6CDI1 A0A1B0FQ19 A0A0L0BMJ6 E9G6L8 A0A0P5FVY6 A0A3R7QMB5 W5JFC1 A0A182IY80 A0A0N8AY16 A0A061QLL8 A0A0P5Q7K3 A0A0P5NU05 A0A182W5G6 A0A1A9XSL1 A0A1B0AQG1 B4PYM0 B3NY63 W8BAI9 B3DN57 B4L8Q4 Q9VRE0 A0A084VLE4 U5EW59 A0A0N8DUS9 A0A1A9W8N8 A0A182RMW2 D6WKL9 A0A0N8EE82 A0A0P5QHP2 A0A026W473 A0A1W4VWQ0 A0A182XXU9 A0A1I8Q0G6 A0A0P6EJG1 B4NCR5 A0A182SYJ8 B4M1T6 A0A1I8MLD4 A0A182UEW2 Q7PS25 A0A182VGX2 B4I6D5 A0A182PBK1 A0A3L8E1R2 B3N0J0 A0A3B0J887 A0A0N0U4L1 A0A0P5IFR8 A0A1W4WSH1 B4JIP5 A0A182GQJ9 A0A087ZWD2 A0A310SNA2 F4W854 A0A195EYT6 A0A158NDQ5 A0A151X418 A0A0M5J0V3 A0A154PSC3 A0A0L7QV25 E2A0N1 A0A2M4A226 A0A034VC64 B5DNT0 A0A2R5LE35 A0A2A3ELE9 A0A293N280 A0A232FJU7 K7JBD1 A0A1B6L1G3 A0A0C9RC03
A0A2H1VSY6 A0A212EGW9 A0A1B6HC36 S4NTV6 A0A067RGL4 A0A2J7PE11 A0A224XKS3 A0A0N7Z980 A0A069DWJ8 R4FP94 A0A0A9YZB0 A0A023F931 A0A1Y1LEM1 A0A1B6DKE0 A0A0K8TMR8 Q17DE3 A0A1Q3F4A6 A0A0A1WZY7 A0A0K8UYU3 A0A034VB92 A0A182HAS4 A0A1A9UEI2 A0A1B0ADN2 A0A0P6CDI1 A0A1B0FQ19 A0A0L0BMJ6 E9G6L8 A0A0P5FVY6 A0A3R7QMB5 W5JFC1 A0A182IY80 A0A0N8AY16 A0A061QLL8 A0A0P5Q7K3 A0A0P5NU05 A0A182W5G6 A0A1A9XSL1 A0A1B0AQG1 B4PYM0 B3NY63 W8BAI9 B3DN57 B4L8Q4 Q9VRE0 A0A084VLE4 U5EW59 A0A0N8DUS9 A0A1A9W8N8 A0A182RMW2 D6WKL9 A0A0N8EE82 A0A0P5QHP2 A0A026W473 A0A1W4VWQ0 A0A182XXU9 A0A1I8Q0G6 A0A0P6EJG1 B4NCR5 A0A182SYJ8 B4M1T6 A0A1I8MLD4 A0A182UEW2 Q7PS25 A0A182VGX2 B4I6D5 A0A182PBK1 A0A3L8E1R2 B3N0J0 A0A3B0J887 A0A0N0U4L1 A0A0P5IFR8 A0A1W4WSH1 B4JIP5 A0A182GQJ9 A0A087ZWD2 A0A310SNA2 F4W854 A0A195EYT6 A0A158NDQ5 A0A151X418 A0A0M5J0V3 A0A154PSC3 A0A0L7QV25 E2A0N1 A0A2M4A226 A0A034VC64 B5DNT0 A0A2R5LE35 A0A2A3ELE9 A0A293N280 A0A232FJU7 K7JBD1 A0A1B6L1G3 A0A0C9RC03
PDB
6FTI
E-value=0,
Score=2642
Ontologies
PATHWAY
GO
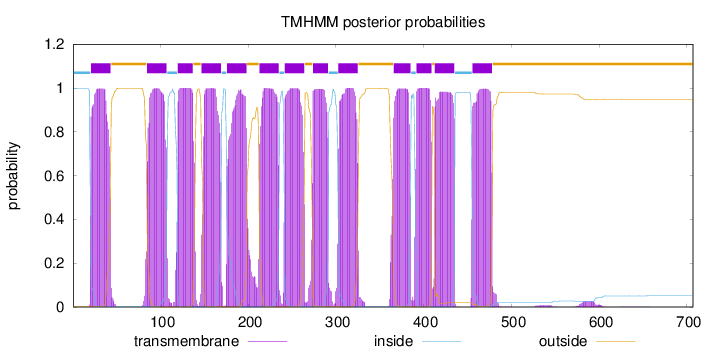
Topology
Length:
707
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
280.60402
Exp number, first 60 AAs:
21.90087
Total prob of N-in:
0.99686
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 84
TMhelix
85 - 107
inside
108 - 119
TMhelix
120 - 137
outside
138 - 146
TMhelix
147 - 169
inside
170 - 175
TMhelix
176 - 198
outside
199 - 212
TMhelix
213 - 235
inside
236 - 241
TMhelix
242 - 264
outside
265 - 273
TMhelix
274 - 291
inside
292 - 302
TMhelix
303 - 325
outside
326 - 365
TMhelix
366 - 385
inside
386 - 391
TMhelix
392 - 409
outside
410 - 412
TMhelix
413 - 435
inside
436 - 455
TMhelix
456 - 478
outside
479 - 707
Population Genetic Test Statistics
Pi
260.091819
Theta
155.641224
Tajima's D
2.439144
CLR
0.326568
CSRT
0.936503174841258
Interpretation
Uncertain
