Gene
KWMTBOMO09287
Pre Gene Modal
BGIBMGA003314
Annotation
PREDICTED:_anoctamin-1_isoform_X2_[Bombyx_mori]
Full name
Anoctamin
Location in the cell
PlasmaMembrane Reliability : 4.493
Sequence
CDS
ATGTCCGACGGAGACCCATCGCTTGAAGAGGACGTACCAAAGCCTCTGCTGGTAAAAGTAGAAGAGAATGTCTTCACGCCTAAGATAACCACTCCGATATACGCCGGTGGTGTTAAACTGCAGCACAAGAGGACCGATCTTGCTGAAACGGAATCCTTCCTCTCAGACTCTCAGAGGAATGACCTTGCATTCCAACAACTCTTCCAGGATGGAGATGATGCATTTCACGAAACGGAGAAGGCGGCTCAAGAGAAGATCGGTGACATCCACGCGATGTCAGTGCTAAGCAACTACCAAGACAACCTCTCCCAAGACATGAATACCGACCGGGACAACGGTAGCGAAGCTTCTACCCGCGCTGCCGAAGAAGTCGGTTGCGGTCAGCCCCAACTAACGAAACCCAATGGCAAACCCGTCGCCAACCATTGCTCAATAGTTATGGGCGGTATCAACAATAACCCCTCGTTGACGTTCAACGATGGCGTTCGTTCGATCGACTACGTGCTGGTCTGGGAGAGCGGTAGGAACGACGCGACCACACCCGAAGCCTACGAGCAGAGGAGGATCTTTGAGCAGAATCTGGAGCACGATGGCTTGCAATTAGAAAGGGAGGAGCCGGAGGGACTTTATGGACTGAATTTTGTTAAGATACACGTGCCGGTCGATGTCCTGAGAGATTACAGTGAGATATTAAAATTAAGGATGCCGATGAAGTTTTCAGCAGTGAAGACTAGTAATTTCATAGATCGAGTTGAGGCTACTTGGGATAAGTTCATATCAAAGATAAGGGTGGATCCGGAGTTGTTCCCTGAAAGAAGACATCGTCTAACCGCTATTTACTCCAAAGACAAAGAATACTTATTCGACACAACATCAGAATGCTTCTGGACACCCTCGATCCGCTCTAGGATAGTTCAATTCATACTGGACAGGAAGAAGTTCTCGAAGCCGGGTGACGATTTCGCATTCGGTATCAACAGACTCATATCAAACTGTACTTATAGCGCGGCCTATCCCTTACACGATGGTGATCTCAAAACACCGGGTTCGATGCGATATTTGCTGTACACGGAATGGGCGGCTGTGTCTAAATGCCTCAAATACCAACCGCTAGACTACGTAAAAGATTATTTCGGTGTTAAAATAGGACTGTATTTTGCTTGGCTCGGTTTCTACACGCACATGTTGATCCCGGCTTCGCTCGTCGGCCTCATCTGTTTCATATATTCGGCCGCGACCGTGTACTCCAACAAACCGAGTGAAGACATTTGCAACTACAACTCGACGATAAAGATGTGTCCGCAGTGCGACTACTTCTGCGAGTTCTGGGATCTGAAGGCGACCTGTCTGCAGTCACGCATCACGTACCTCTTCGACAACTCCACCACCGTATTCTTCGCCATATTCATGAGCTTTTGGGCGGCGTGCTTCCTGGAGCTTTGGAAGCGCTACTCGGCCGAGATGACGCACCGCTGGGACCTCACCGGGTTCGACGTGTACGAGGAGCACCCGCGCCCGCAGTACCTGGCGCGCCTCGCGCACGTCAAGCGACGACAGCTAAACGTGGTGACGGGCGAGAACGAGCCGATGGTCCCGTTCTGGCGCATGCGGCTGCCCGCCACGCTCATGTCGTTCAGCGTGGTGGCGCTGCTGGTGCTGCTGGCGCTGGCCACCGTGCTGGGCGTGGTGCTGTACCGCATGTCGCTGCTGGGCGCCGCCGTGCTGCGCCACAACGACAACGTGCTCATCACGTACAGCCCCATCATGTTCACCACCGCGACGGCCGCCTGCATCAACCTGTGCTTCATCTTCCTCTTCAACTATATCTACCAGTACCTAGCCGAATGGTTGACGGAGAAGGAACTGCTTCGGACGCAGACCGAGTTCGACGACTCGCTCACTCTCAAGATATACCTGCTCCAGTTCGTGAACTACTACGCCTCGATATTCTACATCGCTTTCTTCAAAGGGAAAATCATCGGTCGACCCGGAGAGTATATTAGATTCTTCGGGCACAGACAAGAGGAGTGCGCTCCCGGAGGCTGTCTTCTAGAGCTGTCCATCCAGCTGGCCATAATAATGGTCGGGAAGCAGTTCATCAACACGATCGTGGAGATGATGATGCCGTACCTGCTCTCCTGGTGGAACATCATCAGGACCATCGGCAGGAAGAGACAGTTAAAAAGTCCGCTTCAGTGGGTTAAAGACTTTAAGTTGGTCGACTTCCCGAACATGGGACTGTTTCCTGAATATTTGGAAATGATCCTGCAGTACGGGTTCGTGACGATCTTCGTGGCGGCGTTCCCGCTGGCGCCGCTGTTCGCGCTGATCAACAACGTGCTGGAGATGCGGCTGGACGCGCGCAAGTTCCTCACGTGCTTCCGCCGCCCCGTGCCGCAGCGCGTCACCGACATCGGCGTCTGGTACCGCATCCTCGACTCCATCGGCAAGCTCAGCGTCATCACCAACGGCTTCATAATAGCGTTCACGTCGGAGTTCATCCCGCGGCTGGTGTACGCGTTCACGCACGGCTCGCTCGAGCAGTACGTCAACAACACGCTGGCCGACTTCAACATCACCGTGCTCGAGCACCCGCCGCTCAACTCCACGTACGGGGGGACCATGTGCAGCTTCCCCGGCTACTGGGACTATAAGAACGGCAAGGACTATGCGCACAGCACCTGGTACTGGCACATCATCGGAGCCAGACTCGCCTTCGTCGTCGTCTTCGAGAACGTAGTGGCCCTGGGAACGATAATAGTGAAGTGGGCGATCCCGGACATGTCGGGCGAGCTCCGGGACCGCATCCGGCGCGAGGCCTACGTCATCAACTCGTACATCCTGGAGGAGACTCGCGCGCGCTCGCAGGCCGGGCGTGACTCGCAAGGCGACACTACCGAGCCAAACTGGAACCATATAGCTTCGCTCTCGGGCTCGGACTTTGACCTAGCGGCTCACGGCGACCAAGAGGACCTCGCCGAGCTCCGGGAGCGCGCCGCCGCCGCACGCCCCCCGCCCGACCCCGCGGGCCCCCCGGTGGACCCAGCCGGCCCCCCGCCCGCCCCCGTCTAG
Protein
MSDGDPSLEEDVPKPLLVKVEENVFTPKITTPIYAGGVKLQHKRTDLAETESFLSDSQRNDLAFQQLFQDGDDAFHETEKAAQEKIGDIHAMSVLSNYQDNLSQDMNTDRDNGSEASTRAAEEVGCGQPQLTKPNGKPVANHCSIVMGGINNNPSLTFNDGVRSIDYVLVWESGRNDATTPEAYEQRRIFEQNLEHDGLQLEREEPEGLYGLNFVKIHVPVDVLRDYSEILKLRMPMKFSAVKTSNFIDRVEATWDKFISKIRVDPELFPERRHRLTAIYSKDKEYLFDTTSECFWTPSIRSRIVQFILDRKKFSKPGDDFAFGINRLISNCTYSAAYPLHDGDLKTPGSMRYLLYTEWAAVSKCLKYQPLDYVKDYFGVKIGLYFAWLGFYTHMLIPASLVGLICFIYSAATVYSNKPSEDICNYNSTIKMCPQCDYFCEFWDLKATCLQSRITYLFDNSTTVFFAIFMSFWAACFLELWKRYSAEMTHRWDLTGFDVYEEHPRPQYLARLAHVKRRQLNVVTGENEPMVPFWRMRLPATLMSFSVVALLVLLALATVLGVVLYRMSLLGAAVLRHNDNVLITYSPIMFTTATAACINLCFIFLFNYIYQYLAEWLTEKELLRTQTEFDDSLTLKIYLLQFVNYYASIFYIAFFKGKIIGRPGEYIRFFGHRQEECAPGGCLLELSIQLAIIMVGKQFINTIVEMMMPYLLSWWNIIRTIGRKRQLKSPLQWVKDFKLVDFPNMGLFPEYLEMILQYGFVTIFVAAFPLAPLFALINNVLEMRLDARKFLTCFRRPVPQRVTDIGVWYRILDSIGKLSVITNGFIIAFTSEFIPRLVYAFTHGSLEQYVNNTLADFNITVLEHPPLNSTYGGTMCSFPGYWDYKNGKDYAHSTWYWHIIGARLAFVVVFENVVALGTIIVKWAIPDMSGELRDRIRREAYVINSYILEETRARSQAGRDSQGDTTEPNWNHIASLSGSDFDLAAHGDQEDLAELRERAAAARPPPDPAGPPVDPAGPPPAPV
Summary
Similarity
Belongs to the anoctamin family.
Belongs to the 14-3-3 family.
Belongs to the 14-3-3 family.
Feature
chain Anoctamin
Uniprot
A0A2A4JB80
H9J1C7
A0A194PJV5
A0A3S2NIQ9
A0A2A4JBM3
A0A2H1VAR7
+ More
A0A2W1BTH6 A0A194RKF5 D6WLJ8 A0A139WH80 A0A158NR82 A0A195BFB9 A0A195FIV4 A0A2A3E3P4 F4WRG7 A0A154PEC9 A0A151X4V7 A0A151IU31 Q175J3 E0VLJ2 A0A151IFS5 A0A3L8DK91 A0A026W5Q0 A0A232FE49 K7IMJ8 A0A0M8ZZT9 A0A2J7RT78 A0A336MPV6 A0A336LPF7 A0A336LQ11 A0A336LP51 A0A336L6T4 A0A336LT51 A0A0C9R9J9 A0A1J1IKG9 E2BRL6 A0A1J1IH46 A0A088APQ9 A0A1S4FED6 A0A310SM65 B4PNI3 B3P099 Q9VDV4 Q293M2 A0A1W4W9R4 E9IRX4 B4QTB8 A0A0R1E267 A0A0R1E0N2 Q8IN71 A0A0M4EP19 Q86P24 A0A1I8MZX3 A0A3B0KJ15 A0A0B4JCY1 A0A0R3NKD5 T1PLD3 A0A0L0CJY0 A0A1W4W998 A0A0B4KGH4 A0A0R1E0M9 A0A1W4VXG2 B4N8J7 B4JYY8 A0A1B6EAV7 I5APX8 A0A1I8MZW9 A0A1A9X1M3 W8BBX0 A0A226E445 A0A1I8MZW6 A0A034VVM6 B4LYZ5 E9G415 B3M242 A0A034VUV9 W8B082 A0A1A9XP68 A0A0P5BBJ0 A0A0Q9WGH5 A0A0P5SGR4 A0A0P6GQR2 A0A1A9ZXX6 U4UV79 A0A0P4ZKL5 A0A0N8CC67 A0A034VTW3 A0A0P6CM45 A0A0P5AB55 A0A0P5LMW9 A0A0P5K112 A0A0P6F651 A0A0P5SCH0 A0A0P5NJT5 A0A0P5IEU7 A0A0P6CW00 A0A0P5H052 A0A0P5ZDW7 A0A0N8AQQ9
A0A2W1BTH6 A0A194RKF5 D6WLJ8 A0A139WH80 A0A158NR82 A0A195BFB9 A0A195FIV4 A0A2A3E3P4 F4WRG7 A0A154PEC9 A0A151X4V7 A0A151IU31 Q175J3 E0VLJ2 A0A151IFS5 A0A3L8DK91 A0A026W5Q0 A0A232FE49 K7IMJ8 A0A0M8ZZT9 A0A2J7RT78 A0A336MPV6 A0A336LPF7 A0A336LQ11 A0A336LP51 A0A336L6T4 A0A336LT51 A0A0C9R9J9 A0A1J1IKG9 E2BRL6 A0A1J1IH46 A0A088APQ9 A0A1S4FED6 A0A310SM65 B4PNI3 B3P099 Q9VDV4 Q293M2 A0A1W4W9R4 E9IRX4 B4QTB8 A0A0R1E267 A0A0R1E0N2 Q8IN71 A0A0M4EP19 Q86P24 A0A1I8MZX3 A0A3B0KJ15 A0A0B4JCY1 A0A0R3NKD5 T1PLD3 A0A0L0CJY0 A0A1W4W998 A0A0B4KGH4 A0A0R1E0M9 A0A1W4VXG2 B4N8J7 B4JYY8 A0A1B6EAV7 I5APX8 A0A1I8MZW9 A0A1A9X1M3 W8BBX0 A0A226E445 A0A1I8MZW6 A0A034VVM6 B4LYZ5 E9G415 B3M242 A0A034VUV9 W8B082 A0A1A9XP68 A0A0P5BBJ0 A0A0Q9WGH5 A0A0P5SGR4 A0A0P6GQR2 A0A1A9ZXX6 U4UV79 A0A0P4ZKL5 A0A0N8CC67 A0A034VTW3 A0A0P6CM45 A0A0P5AB55 A0A0P5LMW9 A0A0P5K112 A0A0P6F651 A0A0P5SCH0 A0A0P5NJT5 A0A0P5IEU7 A0A0P6CW00 A0A0P5H052 A0A0P5ZDW7 A0A0N8AQQ9
Pubmed
19121390
26354079
28756777
18362917
19820115
21347285
+ More
21719571 17510324 20566863 30249741 24508170 28648823 20075255 20798317 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665 25315136 26108605 24495485 25348373 21292972 23537049
21719571 17510324 20566863 30249741 24508170 28648823 20075255 20798317 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21282665 25315136 26108605 24495485 25348373 21292972 23537049
EMBL
NWSH01002254
PCG68804.1
BABH01023961
KQ459602
KPI93602.1
RSAL01000026
+ More
RVE52062.1 PCG68803.1 ODYU01001553 SOQ37935.1 KZ149923 PZC77651.1 KQ460106 KPJ17815.1 KQ971343 EFA04121.2 KYB27211.1 ADTU01023819 KQ976504 KYM82887.1 KQ981522 KYN40595.1 KZ288391 PBC26373.1 GL888285 EGI63238.1 KQ434889 KZC10225.1 KQ982540 KYQ55417.1 KQ980970 KYN10920.1 CH477400 EAT41725.1 DS235271 EEB14248.1 KQ977771 KYN00015.1 QOIP01000007 RLU20756.1 KK107390 EZA51402.1 NNAY01000356 OXU28942.1 KQ435789 KOX74465.1 NEVH01000006 PNF44039.1 UFQS01002074 UFQT01002074 SSX13145.1 SSX32584.1 UFQT01000100 SSX19916.1 SSX19915.1 SSX19914.1 SSX13144.1 SSX32583.1 SSX19913.1 GBYB01009597 JAG79364.1 CVRI01000051 CRK99574.1 GL449965 EFN81708.1 CRK99573.1 KQ763867 OAD54669.1 CM000160 EDW96046.1 CH954181 EDV48473.1 AE014297 BT010299 AAF55685.1 AAQ23617.1 CM000070 EAL29192.2 GL765283 EFZ16613.1 CM000364 EDX12320.1 KRK02862.1 KRK02861.1 AAN13804.1 CP012526 ALC47465.1 BT003522 AAO39526.1 OUUW01000008 SPP83738.1 ADV37339.1 KRT01374.1 KA648910 AFP63539.1 JRES01000301 KNC32555.1 AGB96117.1 KRK02860.1 CH964232 EDW81448.2 CH916378 EDV98603.1 GEDC01002255 JAS35043.1 EIM53013.2 GAMC01012092 JAB94463.1 LNIX01000006 OXA52492.1 GAKP01013057 GAKP01013054 JAC45895.1 CH940650 EDW68098.2 GL732531 EFX85887.1 CH902617 EDV43366.2 GAKP01013056 GAKP01013055 JAC45897.1 GAMC01012090 JAB94465.1 GDIP01191068 JAJ32334.1 KRF83641.1 GDIQ01091925 JAL59801.1 GDIQ01031602 JAN63135.1 KB632380 ERL94141.1 GDIP01215223 JAJ08179.1 GDIQ01094123 JAL57603.1 GAKP01013053 JAC45899.1 GDIQ01089481 JAN05256.1 GDIP01205965 JAJ17437.1 GDIQ01171151 JAK80574.1 GDIQ01192340 JAK59385.1 GDIQ01253640 GDIQ01207998 GDIQ01206707 GDIQ01203448 GDIQ01202100 GDIQ01067495 JAN27242.1 GDIQ01097782 GDIQ01097781 JAL53944.1 GDIQ01151514 JAL00212.1 GDIQ01213440 JAK38285.1 GDIQ01089480 GDIQ01084886 JAN05257.1 GDIQ01235695 GDIQ01234544 GDIQ01234543 GDIQ01208000 GDIQ01207997 GDIQ01206709 GDIQ01205147 GDIQ01203449 GDIQ01203447 GDIQ01169357 GDIQ01156955 GDIQ01147975 GDIQ01140012 GDIQ01128527 GDIQ01091924 GDIQ01064643 GDIQ01061388 GDIQ01035749 JAK17182.1 GDIP01045592 LRGB01000446 JAM58123.1 KZS19048.1 GDIQ01252187 JAJ99537.1
RVE52062.1 PCG68803.1 ODYU01001553 SOQ37935.1 KZ149923 PZC77651.1 KQ460106 KPJ17815.1 KQ971343 EFA04121.2 KYB27211.1 ADTU01023819 KQ976504 KYM82887.1 KQ981522 KYN40595.1 KZ288391 PBC26373.1 GL888285 EGI63238.1 KQ434889 KZC10225.1 KQ982540 KYQ55417.1 KQ980970 KYN10920.1 CH477400 EAT41725.1 DS235271 EEB14248.1 KQ977771 KYN00015.1 QOIP01000007 RLU20756.1 KK107390 EZA51402.1 NNAY01000356 OXU28942.1 KQ435789 KOX74465.1 NEVH01000006 PNF44039.1 UFQS01002074 UFQT01002074 SSX13145.1 SSX32584.1 UFQT01000100 SSX19916.1 SSX19915.1 SSX19914.1 SSX13144.1 SSX32583.1 SSX19913.1 GBYB01009597 JAG79364.1 CVRI01000051 CRK99574.1 GL449965 EFN81708.1 CRK99573.1 KQ763867 OAD54669.1 CM000160 EDW96046.1 CH954181 EDV48473.1 AE014297 BT010299 AAF55685.1 AAQ23617.1 CM000070 EAL29192.2 GL765283 EFZ16613.1 CM000364 EDX12320.1 KRK02862.1 KRK02861.1 AAN13804.1 CP012526 ALC47465.1 BT003522 AAO39526.1 OUUW01000008 SPP83738.1 ADV37339.1 KRT01374.1 KA648910 AFP63539.1 JRES01000301 KNC32555.1 AGB96117.1 KRK02860.1 CH964232 EDW81448.2 CH916378 EDV98603.1 GEDC01002255 JAS35043.1 EIM53013.2 GAMC01012092 JAB94463.1 LNIX01000006 OXA52492.1 GAKP01013057 GAKP01013054 JAC45895.1 CH940650 EDW68098.2 GL732531 EFX85887.1 CH902617 EDV43366.2 GAKP01013056 GAKP01013055 JAC45897.1 GAMC01012090 JAB94465.1 GDIP01191068 JAJ32334.1 KRF83641.1 GDIQ01091925 JAL59801.1 GDIQ01031602 JAN63135.1 KB632380 ERL94141.1 GDIP01215223 JAJ08179.1 GDIQ01094123 JAL57603.1 GAKP01013053 JAC45899.1 GDIQ01089481 JAN05256.1 GDIP01205965 JAJ17437.1 GDIQ01171151 JAK80574.1 GDIQ01192340 JAK59385.1 GDIQ01253640 GDIQ01207998 GDIQ01206707 GDIQ01203448 GDIQ01202100 GDIQ01067495 JAN27242.1 GDIQ01097782 GDIQ01097781 JAL53944.1 GDIQ01151514 JAL00212.1 GDIQ01213440 JAK38285.1 GDIQ01089480 GDIQ01084886 JAN05257.1 GDIQ01235695 GDIQ01234544 GDIQ01234543 GDIQ01208000 GDIQ01207997 GDIQ01206709 GDIQ01205147 GDIQ01203449 GDIQ01203447 GDIQ01169357 GDIQ01156955 GDIQ01147975 GDIQ01140012 GDIQ01128527 GDIQ01091924 GDIQ01064643 GDIQ01061388 GDIQ01035749 JAK17182.1 GDIP01045592 LRGB01000446 JAM58123.1 KZS19048.1 GDIQ01252187 JAJ99537.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000283053
UP000053240
UP000007266
+ More
UP000005205 UP000078540 UP000078541 UP000242457 UP000007755 UP000076502 UP000075809 UP000078492 UP000008820 UP000009046 UP000078542 UP000279307 UP000053097 UP000215335 UP000002358 UP000053105 UP000235965 UP000183832 UP000008237 UP000005203 UP000002282 UP000008711 UP000000803 UP000001819 UP000192221 UP000000304 UP000092553 UP000095301 UP000268350 UP000037069 UP000007798 UP000001070 UP000091820 UP000198287 UP000008792 UP000000305 UP000007801 UP000092443 UP000092445 UP000030742 UP000076858
UP000005205 UP000078540 UP000078541 UP000242457 UP000007755 UP000076502 UP000075809 UP000078492 UP000008820 UP000009046 UP000078542 UP000279307 UP000053097 UP000215335 UP000002358 UP000053105 UP000235965 UP000183832 UP000008237 UP000005203 UP000002282 UP000008711 UP000000803 UP000001819 UP000192221 UP000000304 UP000092553 UP000095301 UP000268350 UP000037069 UP000007798 UP000001070 UP000091820 UP000198287 UP000008792 UP000000305 UP000007801 UP000092443 UP000092445 UP000030742 UP000076858
Pfam
Interpro
IPR032394
Anoct_dimer
+ More
IPR007632 Anoctamin
IPR000184 Bac_surfAg_D15
IPR019523 Prot_Pase1_reg-su15A/B_C
IPR004895 Prenylated_rab_accept_PRA1
IPR016024 ARM-type_fold
IPR013930 RNA_pol_II_AP1_N
IPR013929 RNA_pol_II_AP1_C
IPR036815 14-3-3_dom_sf
IPR019775 WD40_repeat_CS
IPR001848 Ribosomal_S10
IPR000308 14-3-3
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR022052 Histone-bd_RBBP4_N
IPR019343 Unchr_KLRAQ/TTKRSYEDQ_N
IPR023409 14-3-3_CS
IPR027486 Ribosomal_S10_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036838 Ribosomal_S10_dom_sf
IPR023410 14-3-3_domain
IPR019348 KLRAQ/TTKRSYEDQ_C
IPR007632 Anoctamin
IPR000184 Bac_surfAg_D15
IPR019523 Prot_Pase1_reg-su15A/B_C
IPR004895 Prenylated_rab_accept_PRA1
IPR016024 ARM-type_fold
IPR013930 RNA_pol_II_AP1_N
IPR013929 RNA_pol_II_AP1_C
IPR036815 14-3-3_dom_sf
IPR019775 WD40_repeat_CS
IPR001848 Ribosomal_S10
IPR000308 14-3-3
IPR036322 WD40_repeat_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR022052 Histone-bd_RBBP4_N
IPR019343 Unchr_KLRAQ/TTKRSYEDQ_N
IPR023409 14-3-3_CS
IPR027486 Ribosomal_S10_dom
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036838 Ribosomal_S10_dom_sf
IPR023410 14-3-3_domain
IPR019348 KLRAQ/TTKRSYEDQ_C
Gene 3D
ProteinModelPortal
A0A2A4JB80
H9J1C7
A0A194PJV5
A0A3S2NIQ9
A0A2A4JBM3
A0A2H1VAR7
+ More
A0A2W1BTH6 A0A194RKF5 D6WLJ8 A0A139WH80 A0A158NR82 A0A195BFB9 A0A195FIV4 A0A2A3E3P4 F4WRG7 A0A154PEC9 A0A151X4V7 A0A151IU31 Q175J3 E0VLJ2 A0A151IFS5 A0A3L8DK91 A0A026W5Q0 A0A232FE49 K7IMJ8 A0A0M8ZZT9 A0A2J7RT78 A0A336MPV6 A0A336LPF7 A0A336LQ11 A0A336LP51 A0A336L6T4 A0A336LT51 A0A0C9R9J9 A0A1J1IKG9 E2BRL6 A0A1J1IH46 A0A088APQ9 A0A1S4FED6 A0A310SM65 B4PNI3 B3P099 Q9VDV4 Q293M2 A0A1W4W9R4 E9IRX4 B4QTB8 A0A0R1E267 A0A0R1E0N2 Q8IN71 A0A0M4EP19 Q86P24 A0A1I8MZX3 A0A3B0KJ15 A0A0B4JCY1 A0A0R3NKD5 T1PLD3 A0A0L0CJY0 A0A1W4W998 A0A0B4KGH4 A0A0R1E0M9 A0A1W4VXG2 B4N8J7 B4JYY8 A0A1B6EAV7 I5APX8 A0A1I8MZW9 A0A1A9X1M3 W8BBX0 A0A226E445 A0A1I8MZW6 A0A034VVM6 B4LYZ5 E9G415 B3M242 A0A034VUV9 W8B082 A0A1A9XP68 A0A0P5BBJ0 A0A0Q9WGH5 A0A0P5SGR4 A0A0P6GQR2 A0A1A9ZXX6 U4UV79 A0A0P4ZKL5 A0A0N8CC67 A0A034VTW3 A0A0P6CM45 A0A0P5AB55 A0A0P5LMW9 A0A0P5K112 A0A0P6F651 A0A0P5SCH0 A0A0P5NJT5 A0A0P5IEU7 A0A0P6CW00 A0A0P5H052 A0A0P5ZDW7 A0A0N8AQQ9
A0A2W1BTH6 A0A194RKF5 D6WLJ8 A0A139WH80 A0A158NR82 A0A195BFB9 A0A195FIV4 A0A2A3E3P4 F4WRG7 A0A154PEC9 A0A151X4V7 A0A151IU31 Q175J3 E0VLJ2 A0A151IFS5 A0A3L8DK91 A0A026W5Q0 A0A232FE49 K7IMJ8 A0A0M8ZZT9 A0A2J7RT78 A0A336MPV6 A0A336LPF7 A0A336LQ11 A0A336LP51 A0A336L6T4 A0A336LT51 A0A0C9R9J9 A0A1J1IKG9 E2BRL6 A0A1J1IH46 A0A088APQ9 A0A1S4FED6 A0A310SM65 B4PNI3 B3P099 Q9VDV4 Q293M2 A0A1W4W9R4 E9IRX4 B4QTB8 A0A0R1E267 A0A0R1E0N2 Q8IN71 A0A0M4EP19 Q86P24 A0A1I8MZX3 A0A3B0KJ15 A0A0B4JCY1 A0A0R3NKD5 T1PLD3 A0A0L0CJY0 A0A1W4W998 A0A0B4KGH4 A0A0R1E0M9 A0A1W4VXG2 B4N8J7 B4JYY8 A0A1B6EAV7 I5APX8 A0A1I8MZW9 A0A1A9X1M3 W8BBX0 A0A226E445 A0A1I8MZW6 A0A034VVM6 B4LYZ5 E9G415 B3M242 A0A034VUV9 W8B082 A0A1A9XP68 A0A0P5BBJ0 A0A0Q9WGH5 A0A0P5SGR4 A0A0P6GQR2 A0A1A9ZXX6 U4UV79 A0A0P4ZKL5 A0A0N8CC67 A0A034VTW3 A0A0P6CM45 A0A0P5AB55 A0A0P5LMW9 A0A0P5K112 A0A0P6F651 A0A0P5SCH0 A0A0P5NJT5 A0A0P5IEU7 A0A0P6CW00 A0A0P5H052 A0A0P5ZDW7 A0A0N8AQQ9
PDB
5OYG
E-value=8.88505e-170,
Score=1536
Ontologies
GO
PANTHER
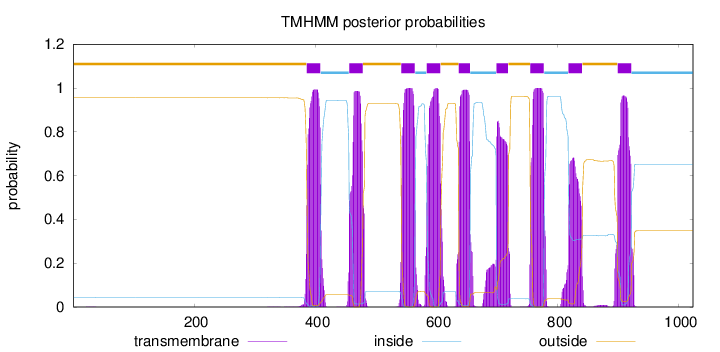
Topology
Subcellular location
Membrane
Nucleus
Nucleus
Length:
1023
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
191.43648
Exp number, first 60 AAs:
0.00361
Total prob of N-in:
0.04414
outside
1 - 385
TMhelix
386 - 408
inside
409 - 455
TMhelix
456 - 478
outside
479 - 541
TMhelix
542 - 564
inside
565 - 583
TMhelix
584 - 606
outside
607 - 636
TMhelix
637 - 655
inside
656 - 698
TMhelix
699 - 718
outside
719 - 754
TMhelix
755 - 777
inside
778 - 817
TMhelix
818 - 840
outside
841 - 898
TMhelix
899 - 921
inside
922 - 1023
Population Genetic Test Statistics
Pi
216.521007
Theta
162.344276
Tajima's D
1.010324
CLR
0.442365
CSRT
0.660666966651667
Interpretation
Uncertain
