Gene
KWMTBOMO09285
Pre Gene Modal
BGIBMGA003315
Annotation
PREDICTED:_adenylate_cyclase_type_8_[Bombyx_mori]
Full name
Adenylate cyclase
Location in the cell
PlasmaMembrane Reliability : 4.2
Sequence
CDS
ATGGTTCACAAAATAGTCTGGAGTGTGGCCTTCGCGTGGCTGAACATCGGGGTCTGTCTGCTCGGATGCTGGCGGTGCTTCGCCAACAACTACCTGCACTGGGCAGCAGCGGCTACTTGGATACTGTTAATTGCTCAAGGTATAAGCGGACAGGGAATCGGCTTCCAGACTCCTCAAAACCAGGTCTGGTATATGCTGTTCGTGGTGTTCGTTCCCTATGCGATGCTTCCGCTCTCACTCGTGTGGTGCATCGTGATTGGAATTCTATCGTTTCTGTCTCACATCCTCGCGACGGCTGTGGACATAGAGAACCTGGAACAAAATAACATGATTCAGGCGGCCAGTTCCTGCAGTATCCGTCTACTAACCGGCAATGCGCTTCTATACATTGCTGTAAACTTTGCGGGTATGTACGCCAAATCACTAGCTGATTGGGGCCAGAGAAAGGCCTTTCTGGAGACTCATCGCTCGATGGTCACCAGACAACGAACTAAAACAGAGAGCGACAAGCAATGGAACTTATTTCAGAGCGTAATCCCAGACTTCTTAGCCAAGGAGATCTACAGCCATGTTTCAAAGGTGAAAGGGGATTTCCAAGAGCAGCAATTCAACAATCTTTACATACAGAGACATGAAAACGTGTCCATACTCTACGCAGACATAAAGGGTTTTACTGAGCTATCAAGTAAGTGCAACGCTCAAGAATTGGTCAAATTATTGAATGAATTGTTCGCTAGATTCGACAAATTGGCTTCGGAGAATCATTGTCTCCGAATCAAGTTGCTCGGTGATTGCTATTTCTGCGTGAGTGGTCTGACGGAGCAACGCGCCTCACATGCGTACGATTGCGTCAACATGGGCCTGCATATGATCCGGGCTATCCGTGACGTCCGTTACAATACACAGGTGGACCTGGATATGCGTATCGGCATCCATAGCGGCACGGTGCTGTGCGGGGTGCTAGGTCTGCTCAAGTGGCAGTTCGACCTTTGGTCCTATGACGTCACACTCGCCAACCACATGGAGAGCGGGGGCTTGCCCGGACGAGTGCACATTTCGTCAGCAACATATGAGTGCCTGTGCGGTGCTTTCGAGGTGGAACCTGGTGACGGAGCCTCCAGAGATCCTTACATCAAAGAACAGAACATCACCACTTACCTGATAAAAGCGACGGAGCAGCCTCGGAGAGCCAGGCACCCTTCTCGCAACGGTTTTCCCTTAAGCCAGCAGATCTGGACGACAATGACGCGTCGTCCAAGCGGCAGCATCGATGACGAAACCACGACCGACTGGACCCCGGAAATGCCGTTTCAGAACTATTTAACTGGTGGTACAACTACAAGGGCACGGTTGGACATGTTCCGAGTGGGTCAAGAAGCCGAGGTGGAGGAGGACGCCCCAGTCTCAGATGTTGAAGAAGGCGATATTATTAATCACTCGATAGAGGTGGAAAGCAATCGCCGGATGCGCGTGGACAACATGCGCGCGGTCTCGCTGCGGTTCAGACGGGCGTCCCTCGAACGAAGCTTTGGAGAGCTGGACGAGCGGACCTTCAAGTCTAACGTGCTCTGCTGCTTCGTACTCTGGCTCTTCAATGTAGCCGTCCAGGCGGTCATACATTACAATTGCTGGATCCTGATGATAATATTGGCGAACATGACAATCCCGCTGGTGATATCATTCGTGCTGGTGATGATGGAAGAATTCCCGAGACTGTCGCCGCGACTGGCAAAGTTTTCGGCTGCTCTAAGTGCGACCACGCTCAGGAGAAACATACATATTTGCTGTTTCATCATGATTATGTCTGTGTCCAGCACAATAAAACTGTACGTTTGTCCGTCATCTTTTCCGCCGACTTCGTACAATGGAAGCTACACGTTAAACACCACGTTCTGTGAGGATACAAACTCGACGATGTGCGCGGTCGAAGCCGGCGAATGCTATCGTCCGGAATACGTGGTTTACACGTGGGTCTTGTGCCTGGTCGCTCTGACGTCAGTACTGAAACTCCACTACCAGATCAAAACGTTACTGGCCATCATAGACGTTCTCATGTTCTGCGTTTTGCTCTTGAAGAACTACGACGTTTATTATTTCGTCACGAACGAGGGTGCTACAAATCAACTGCCAGCGTATGTCCAAATGTTAATACTAATGGCAATGTTCCTCGCCGTTGTCATTTACCACGGGCGGCTGGTGGAAGTGACGTCACGTCTCGACTTTCTCTGGAAACTTCAAGCGGAGACCGATCTCGGAGAAATGGAGGACACACAGAGAATCAACAGACATCTACTGAAGCACATATTACCTGACCATGTCGTAACACACTTCTTGAGTAAAGATCGGTGTCCTAATGAACTGTACTCGCAGTGGCGCGACGAGGTGGGAGTGATGTTCGCCGGCATCCCGAACTTCCACGAGTTCTACTCGGAGACGAAGGCCGTCGACTGCATGCGGTTGCTCAACGAGATAATTATTGATTTCGACAAGCTGTTGATGGAGGAGCGGTTCAAGAGCATCGAGAAGATCAAGACCATCGGAGCCACTTACATGGCCGCCAGTGGACTGGACCCCGATGTTAAGTCTGGTGCTGACGAGTGCGCGCACCTGTGTGCGCTCGTGGACTACGCGTTTGCGCTGCGCGAGGCTCTGGATGACGTCAACAAGCACAGCTTCAACAAATTTCGCCTCAGGGTCGGTATCAGCTGTGGACCTTTGGTGGGCGGCGTAATCGGTGCTCTGAAACCGGTCTACGACATCTGGGGGAACACTGTGAACGAAGCTTCGAGAATGGAGAGCACAGGCGCTATGGACAAGATACAAGTCACCAAATATACTAAACAGTTGTTAGAGGTTCGCGGATACGGCGTGGAGCGCCGTGGTTGCGTCGAGGTGAAGGGCAAGGGGCGCATGGAGACGTGGTGGGTGGTGTCGAGGCGGGGCGCCCCGCGTCCCCCGCGCCGCCCCGCGCCCCACCCCCGCTCCCTCGCCGCGCTCGTCTACACTATGCTGCAGGCGAGAAAGCGCATCTACACGCACCCTCTAGATGCCACGCATATCGTGAATCGAAGCGACAGCATGCGGCCGACCCGCGTGGACACTGCGCCCAGCGGCTCCAACCGGCGCCGCTCAGAGCTGCGGCGGGCGCGCACGCGGCACAACGACCGCCATGCGGACACGGAGCTGAGGGCGTTGGGTCGTCATTCCGCGTCGGCGCCGCACACTCCGACGGCGACGCACTGCCCGCCGCCCCCCGACCCCCCGCAACTGTCCACCATCACCAGGATAGCCCGGTTGAAAGCTAACAGCTTCACATTCCGGTCGCGGCCCAGAGATAAGTCTCCCAAGATCCGGGAGCAGCCTAACGAGGGATCCGGTACATCACTAGCGAACGGAACGCCTGGCTCGTTCCGCATCAGGTCGGTGACGACAGCGTCGTTCTTCAGAAGCCGAAACAAAGACAAGAGGAGCAAGGAGCAAATAGATACTGAAACCAGGAGTGAAGGAGTGGAAAACGGTGACGCCTACACCACCAAAATGTAA
Protein
MVHKIVWSVAFAWLNIGVCLLGCWRCFANNYLHWAAAATWILLIAQGISGQGIGFQTPQNQVWYMLFVVFVPYAMLPLSLVWCIVIGILSFLSHILATAVDIENLEQNNMIQAASSCSIRLLTGNALLYIAVNFAGMYAKSLADWGQRKAFLETHRSMVTRQRTKTESDKQWNLFQSVIPDFLAKEIYSHVSKVKGDFQEQQFNNLYIQRHENVSILYADIKGFTELSSKCNAQELVKLLNELFARFDKLASENHCLRIKLLGDCYFCVSGLTEQRASHAYDCVNMGLHMIRAIRDVRYNTQVDLDMRIGIHSGTVLCGVLGLLKWQFDLWSYDVTLANHMESGGLPGRVHISSATYECLCGAFEVEPGDGASRDPYIKEQNITTYLIKATEQPRRARHPSRNGFPLSQQIWTTMTRRPSGSIDDETTTDWTPEMPFQNYLTGGTTTRARLDMFRVGQEAEVEEDAPVSDVEEGDIINHSIEVESNRRMRVDNMRAVSLRFRRASLERSFGELDERTFKSNVLCCFVLWLFNVAVQAVIHYNCWILMIILANMTIPLVISFVLVMMEEFPRLSPRLAKFSAALSATTLRRNIHICCFIMIMSVSSTIKLYVCPSSFPPTSYNGSYTLNTTFCEDTNSTMCAVEAGECYRPEYVVYTWVLCLVALTSVLKLHYQIKTLLAIIDVLMFCVLLLKNYDVYYFVTNEGATNQLPAYVQMLILMAMFLAVVIYHGRLVEVTSRLDFLWKLQAETDLGEMEDTQRINRHLLKHILPDHVVTHFLSKDRCPNELYSQWRDEVGVMFAGIPNFHEFYSETKAVDCMRLLNEIIIDFDKLLMEERFKSIEKIKTIGATYMAASGLDPDVKSGADECAHLCALVDYAFALREALDDVNKHSFNKFRLRVGISCGPLVGGVIGALKPVYDIWGNTVNEASRMESTGAMDKIQVTKYTKQLLEVRGYGVERRGCVEVKGKGRMETWWVVSRRGAPRPPRRPAPHPRSLAALVYTMLQARKRIYTHPLDATHIVNRSDSMRPTRVDTAPSGSNRRRSELRRARTRHNDRHADTELRALGRHSASAPHTPTATHCPPPPDPPQLSTITRIARLKANSFTFRSRPRDKSPKIREQPNEGSGTSLANGTPGSFRIRSVTTASFFRSRNKDKRSKEQIDTETRSEGVENGDAYTTKM
Summary
Description
Catalyzes the formation of the signaling molecule cAMP in response to G-protein signaling.
Catalytic Activity
ATP = 3',5'-cyclic AMP + diphosphate
Cofactor
Mg(2+)
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Feature
chain Adenylate cyclase
Uniprot
H9J1C8
A0A2H1VD99
A0A437BNT1
A0A212F546
A0A194RK59
A0A194PJK1
+ More
A0A2W1BWS2 A0A2A4JYB9 A0A2A4J4W6 A0A2H1VNT2 A0A0L7KXB4 A0A2A4J4U7 A0A0N0BIT4 A0A154P206 J9JRH3 A0A2S2NEH7 A0A2S2Q9X8 A0A2H8TUF5 T1IKH0 A0A2R5L7Z5 A0A131XVB0 A0A1J1IIY5 A0A1L8FTY2 A0A1L8G048 A4IGT8 A0A151P8F6 A0A151P7X1 A0A2I0MW10 A0A1V4KVB2 A0A384BYD8 D2H7A3 U3JIS7 A0A1D5PNE8 A0A0Q3MQF7 A0A452VC85 W5N4M7 A0A3Q7VGE3 A0A452HUQ3 A0A1W5AC82 A0A2D4Q209 A0A0F7Z1G0 A0A2D4FAP3 A0A3B3CNM2 A0A1S2ZQ21 G1KKD9 F7BCI5 A0A3B4TKY1 I3K3B0 A0A3P8R1Q8 A0A226N9N4 A0A3B3S1H4 A0A098M1V3 A0A401RWJ0 A0A0B8RRT3 A0A139WI81 A0A452G620 A0A1W7RJN6 T1DP06 A0A452G691 E1BQ12 A0A0F7ZE02 A0A287AWF4 D6WK22 J3RYC9 L8IAT5 A0A2K5R7M0 A0A384ANN7 A0A287DAM6 E2RCX6 F6R859 A0A340WM11 A0A3Q7S0N3 A0A287DCD9 H0V9J2 A0A3Q7QGH2 A0A2K5EK24 A0A2U3WLQ4 U3DX42 A0A2D0T7E1 A0A3P9Q9A5 A0A1S3LVG8 G5B4Q6 U3DLD7 A0A3B5QDJ9 A0A3Q0GSN8 A0A2I2ZTJ4 G1R0Z0 A0A2U4B351 A0A096N068 A0A2K5M765 H9F9J8 A0A1D5Q874 G7PCU6 B7UCF8 A0A1S3FW47 A0A2K6DDC0 A0A2K5XPW5 A0A2Y9DED6 H2PR79
A0A2W1BWS2 A0A2A4JYB9 A0A2A4J4W6 A0A2H1VNT2 A0A0L7KXB4 A0A2A4J4U7 A0A0N0BIT4 A0A154P206 J9JRH3 A0A2S2NEH7 A0A2S2Q9X8 A0A2H8TUF5 T1IKH0 A0A2R5L7Z5 A0A131XVB0 A0A1J1IIY5 A0A1L8FTY2 A0A1L8G048 A4IGT8 A0A151P8F6 A0A151P7X1 A0A2I0MW10 A0A1V4KVB2 A0A384BYD8 D2H7A3 U3JIS7 A0A1D5PNE8 A0A0Q3MQF7 A0A452VC85 W5N4M7 A0A3Q7VGE3 A0A452HUQ3 A0A1W5AC82 A0A2D4Q209 A0A0F7Z1G0 A0A2D4FAP3 A0A3B3CNM2 A0A1S2ZQ21 G1KKD9 F7BCI5 A0A3B4TKY1 I3K3B0 A0A3P8R1Q8 A0A226N9N4 A0A3B3S1H4 A0A098M1V3 A0A401RWJ0 A0A0B8RRT3 A0A139WI81 A0A452G620 A0A1W7RJN6 T1DP06 A0A452G691 E1BQ12 A0A0F7ZE02 A0A287AWF4 D6WK22 J3RYC9 L8IAT5 A0A2K5R7M0 A0A384ANN7 A0A287DAM6 E2RCX6 F6R859 A0A340WM11 A0A3Q7S0N3 A0A287DCD9 H0V9J2 A0A3Q7QGH2 A0A2K5EK24 A0A2U3WLQ4 U3DX42 A0A2D0T7E1 A0A3P9Q9A5 A0A1S3LVG8 G5B4Q6 U3DLD7 A0A3B5QDJ9 A0A3Q0GSN8 A0A2I2ZTJ4 G1R0Z0 A0A2U4B351 A0A096N068 A0A2K5M765 H9F9J8 A0A1D5Q874 G7PCU6 B7UCF8 A0A1S3FW47 A0A2K6DDC0 A0A2K5XPW5 A0A2Y9DED6 H2PR79
EC Number
4.6.1.1
Pubmed
19121390
22118469
26354079
28756777
26227816
27762356
+ More
22293439 23371554 20010809 15592404 24813606 28562605 29451363 17495919 25186727 29240929 30297745 25476704 18362917 19820115 26358130 23758969 25727380 19393038 30723633 23025625 22751099 16341006 19892987 21993624 25243066 21993625 23542700 22398555 25319552 17431167 22002653
22293439 23371554 20010809 15592404 24813606 28562605 29451363 17495919 25186727 29240929 30297745 25476704 18362917 19820115 26358130 23758969 25727380 19393038 30723633 23025625 22751099 16341006 19892987 21993624 25243066 21993625 23542700 22398555 25319552 17431167 22002653
EMBL
BABH01023949
BABH01023950
BABH01023951
ODYU01001670
SOQ38214.1
RSAL01000026
+ More
RVE52061.1 AGBW02010254 OWR48857.1 KQ460106 KPJ17814.1 KQ459602 KPI93601.1 KZ149923 PZC77647.1 NWSH01000401 PCG76668.1 NWSH01003368 PCG66442.1 ODYU01003556 SOQ42473.1 JTDY01004844 KOB67681.1 PCG66443.1 KQ435726 KOX78138.1 KQ434786 KZC05160.1 ABLF02010787 ABLF02010788 ABLF02033700 ABLF02033702 ABLF02033705 ABLF02033707 ABLF02033708 ABLF02049221 ABLF02057914 GGMR01002974 MBY15593.1 GGMS01005147 MBY74350.1 GFXV01005636 MBW17441.1 JH430530 GGLE01001497 MBY05623.1 GEFM01004563 JAP71233.1 CVRI01000051 CRK99500.1 CM004477 OCT75025.1 CM004476 OCT77174.1 BC135243 AAI35244.1 AKHW03000629 KYO45189.1 KYO45191.1 AKCR02000001 PKK33863.1 LSYS01001531 OPJ88376.1 ACTA01113176 ACTA01121176 ACTA01129176 ACTA01137176 ACTA01145175 ACTA01153174 GL192550 EFB24884.1 AGTO01000306 AADN05000663 LMAW01000991 KQK84735.1 AHAT01002480 AHAT01002481 AHAT01002482 IACN01108799 LAB64244.1 GBEW01000039 JAI10326.1 IACJ01068186 LAA44541.1 AERX01029748 AERX01029749 AERX01029750 AERX01029751 AERX01029752 AERX01029753 AERX01029754 AERX01029755 AERX01029756 MCFN01000128 OXB64197.1 GBSI01000169 JAC96326.1 BEZZ01000012 GCC22491.1 GBSH01000215 JAG68809.1 KQ971342 KYB27619.1 LWLT01000011 GDAY02000074 JAV51339.1 GAAZ01000082 GBKC01000119 JAA97861.1 JAG45951.1 GBEX01000113 JAI14447.1 AEMK02000022 DQIR01184664 HDB40141.1 EFA03629.2 JU173823 AFJ49349.1 JH881600 ELR53323.1 AGTP01001445 AGTP01001446 AGTP01001447 AGTP01001448 AGTP01001449 AAEX03008817 AAEX03008818 AAKN02041798 AAKN02041799 GAMS01008821 JAB14315.1 JH168480 EHB04267.1 GAMR01003168 GAMP01008056 JAB30764.1 JAB44699.1 CABD030061129 CABD030061130 CABD030061131 ADFV01168929 ADFV01168930 ADFV01168931 ADFV01168932 ADFV01168933 ADFV01168934 ADFV01168935 ADFV01168936 ADFV01168937 ADFV01168938 AHZZ02034445 AHZZ02034446 AHZZ02034447 AHZZ02034448 JU327551 AFE71307.1 JSUE03042472 JSUE03042473 JSUE03042474 AQIA01069298 AQIA01069299 AQIA01069300 AQIA01069301 CM001283 EHH64433.1 FJ472834 ACK57564.1 ABGA01133391 ABGA01133392 ABGA01133393 ABGA01133394 ABGA01160142 ABGA01160143 ABGA01160144 ABGA01293229 ABGA01293230 ABGA01293231
RVE52061.1 AGBW02010254 OWR48857.1 KQ460106 KPJ17814.1 KQ459602 KPI93601.1 KZ149923 PZC77647.1 NWSH01000401 PCG76668.1 NWSH01003368 PCG66442.1 ODYU01003556 SOQ42473.1 JTDY01004844 KOB67681.1 PCG66443.1 KQ435726 KOX78138.1 KQ434786 KZC05160.1 ABLF02010787 ABLF02010788 ABLF02033700 ABLF02033702 ABLF02033705 ABLF02033707 ABLF02033708 ABLF02049221 ABLF02057914 GGMR01002974 MBY15593.1 GGMS01005147 MBY74350.1 GFXV01005636 MBW17441.1 JH430530 GGLE01001497 MBY05623.1 GEFM01004563 JAP71233.1 CVRI01000051 CRK99500.1 CM004477 OCT75025.1 CM004476 OCT77174.1 BC135243 AAI35244.1 AKHW03000629 KYO45189.1 KYO45191.1 AKCR02000001 PKK33863.1 LSYS01001531 OPJ88376.1 ACTA01113176 ACTA01121176 ACTA01129176 ACTA01137176 ACTA01145175 ACTA01153174 GL192550 EFB24884.1 AGTO01000306 AADN05000663 LMAW01000991 KQK84735.1 AHAT01002480 AHAT01002481 AHAT01002482 IACN01108799 LAB64244.1 GBEW01000039 JAI10326.1 IACJ01068186 LAA44541.1 AERX01029748 AERX01029749 AERX01029750 AERX01029751 AERX01029752 AERX01029753 AERX01029754 AERX01029755 AERX01029756 MCFN01000128 OXB64197.1 GBSI01000169 JAC96326.1 BEZZ01000012 GCC22491.1 GBSH01000215 JAG68809.1 KQ971342 KYB27619.1 LWLT01000011 GDAY02000074 JAV51339.1 GAAZ01000082 GBKC01000119 JAA97861.1 JAG45951.1 GBEX01000113 JAI14447.1 AEMK02000022 DQIR01184664 HDB40141.1 EFA03629.2 JU173823 AFJ49349.1 JH881600 ELR53323.1 AGTP01001445 AGTP01001446 AGTP01001447 AGTP01001448 AGTP01001449 AAEX03008817 AAEX03008818 AAKN02041798 AAKN02041799 GAMS01008821 JAB14315.1 JH168480 EHB04267.1 GAMR01003168 GAMP01008056 JAB30764.1 JAB44699.1 CABD030061129 CABD030061130 CABD030061131 ADFV01168929 ADFV01168930 ADFV01168931 ADFV01168932 ADFV01168933 ADFV01168934 ADFV01168935 ADFV01168936 ADFV01168937 ADFV01168938 AHZZ02034445 AHZZ02034446 AHZZ02034447 AHZZ02034448 JU327551 AFE71307.1 JSUE03042472 JSUE03042473 JSUE03042474 AQIA01069298 AQIA01069299 AQIA01069300 AQIA01069301 CM001283 EHH64433.1 FJ472834 ACK57564.1 ABGA01133391 ABGA01133392 ABGA01133393 ABGA01133394 ABGA01160142 ABGA01160143 ABGA01160144 ABGA01293229 ABGA01293230 ABGA01293231
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000037510 UP000053105 UP000076502 UP000007819 UP000183832 UP000186698 UP000050525 UP000053872 UP000190648 UP000261680 UP000008912 UP000016665 UP000000539 UP000051836 UP000291021 UP000018468 UP000286642 UP000291020 UP000192224 UP000261560 UP000079721 UP000001646 UP000002280 UP000261420 UP000005207 UP000265100 UP000198323 UP000261540 UP000287033 UP000007266 UP000291000 UP000009136 UP000008227 UP000233040 UP000261681 UP000005215 UP000002254 UP000002281 UP000265300 UP000286640 UP000005447 UP000286641 UP000233020 UP000245340 UP000221080 UP000242638 UP000087266 UP000006813 UP000008225 UP000002852 UP000189705 UP000001519 UP000001073 UP000245320 UP000028761 UP000233060 UP000006718 UP000009130 UP000233100 UP000081671 UP000233120 UP000233140 UP000248480 UP000001595
UP000037510 UP000053105 UP000076502 UP000007819 UP000183832 UP000186698 UP000050525 UP000053872 UP000190648 UP000261680 UP000008912 UP000016665 UP000000539 UP000051836 UP000291021 UP000018468 UP000286642 UP000291020 UP000192224 UP000261560 UP000079721 UP000001646 UP000002280 UP000261420 UP000005207 UP000265100 UP000198323 UP000261540 UP000287033 UP000007266 UP000291000 UP000009136 UP000008227 UP000233040 UP000261681 UP000005215 UP000002254 UP000002281 UP000265300 UP000286640 UP000005447 UP000286641 UP000233020 UP000245340 UP000221080 UP000242638 UP000087266 UP000006813 UP000008225 UP000002852 UP000189705 UP000001519 UP000001073 UP000245320 UP000028761 UP000233060 UP000006718 UP000009130 UP000233100 UP000081671 UP000233120 UP000233140 UP000248480 UP000001595
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J1C8
A0A2H1VD99
A0A437BNT1
A0A212F546
A0A194RK59
A0A194PJK1
+ More
A0A2W1BWS2 A0A2A4JYB9 A0A2A4J4W6 A0A2H1VNT2 A0A0L7KXB4 A0A2A4J4U7 A0A0N0BIT4 A0A154P206 J9JRH3 A0A2S2NEH7 A0A2S2Q9X8 A0A2H8TUF5 T1IKH0 A0A2R5L7Z5 A0A131XVB0 A0A1J1IIY5 A0A1L8FTY2 A0A1L8G048 A4IGT8 A0A151P8F6 A0A151P7X1 A0A2I0MW10 A0A1V4KVB2 A0A384BYD8 D2H7A3 U3JIS7 A0A1D5PNE8 A0A0Q3MQF7 A0A452VC85 W5N4M7 A0A3Q7VGE3 A0A452HUQ3 A0A1W5AC82 A0A2D4Q209 A0A0F7Z1G0 A0A2D4FAP3 A0A3B3CNM2 A0A1S2ZQ21 G1KKD9 F7BCI5 A0A3B4TKY1 I3K3B0 A0A3P8R1Q8 A0A226N9N4 A0A3B3S1H4 A0A098M1V3 A0A401RWJ0 A0A0B8RRT3 A0A139WI81 A0A452G620 A0A1W7RJN6 T1DP06 A0A452G691 E1BQ12 A0A0F7ZE02 A0A287AWF4 D6WK22 J3RYC9 L8IAT5 A0A2K5R7M0 A0A384ANN7 A0A287DAM6 E2RCX6 F6R859 A0A340WM11 A0A3Q7S0N3 A0A287DCD9 H0V9J2 A0A3Q7QGH2 A0A2K5EK24 A0A2U3WLQ4 U3DX42 A0A2D0T7E1 A0A3P9Q9A5 A0A1S3LVG8 G5B4Q6 U3DLD7 A0A3B5QDJ9 A0A3Q0GSN8 A0A2I2ZTJ4 G1R0Z0 A0A2U4B351 A0A096N068 A0A2K5M765 H9F9J8 A0A1D5Q874 G7PCU6 B7UCF8 A0A1S3FW47 A0A2K6DDC0 A0A2K5XPW5 A0A2Y9DED6 H2PR79
A0A2W1BWS2 A0A2A4JYB9 A0A2A4J4W6 A0A2H1VNT2 A0A0L7KXB4 A0A2A4J4U7 A0A0N0BIT4 A0A154P206 J9JRH3 A0A2S2NEH7 A0A2S2Q9X8 A0A2H8TUF5 T1IKH0 A0A2R5L7Z5 A0A131XVB0 A0A1J1IIY5 A0A1L8FTY2 A0A1L8G048 A4IGT8 A0A151P8F6 A0A151P7X1 A0A2I0MW10 A0A1V4KVB2 A0A384BYD8 D2H7A3 U3JIS7 A0A1D5PNE8 A0A0Q3MQF7 A0A452VC85 W5N4M7 A0A3Q7VGE3 A0A452HUQ3 A0A1W5AC82 A0A2D4Q209 A0A0F7Z1G0 A0A2D4FAP3 A0A3B3CNM2 A0A1S2ZQ21 G1KKD9 F7BCI5 A0A3B4TKY1 I3K3B0 A0A3P8R1Q8 A0A226N9N4 A0A3B3S1H4 A0A098M1V3 A0A401RWJ0 A0A0B8RRT3 A0A139WI81 A0A452G620 A0A1W7RJN6 T1DP06 A0A452G691 E1BQ12 A0A0F7ZE02 A0A287AWF4 D6WK22 J3RYC9 L8IAT5 A0A2K5R7M0 A0A384ANN7 A0A287DAM6 E2RCX6 F6R859 A0A340WM11 A0A3Q7S0N3 A0A287DCD9 H0V9J2 A0A3Q7QGH2 A0A2K5EK24 A0A2U3WLQ4 U3DX42 A0A2D0T7E1 A0A3P9Q9A5 A0A1S3LVG8 G5B4Q6 U3DLD7 A0A3B5QDJ9 A0A3Q0GSN8 A0A2I2ZTJ4 G1R0Z0 A0A2U4B351 A0A096N068 A0A2K5M765 H9F9J8 A0A1D5Q874 G7PCU6 B7UCF8 A0A1S3FW47 A0A2K6DDC0 A0A2K5XPW5 A0A2Y9DED6 H2PR79
PDB
3MAA
E-value=8.41548e-74,
Score=709
Ontologies
PATHWAY
GO
GO:0004016
GO:0000166
GO:0035556
GO:0006171
GO:0016021
GO:0005886
GO:0046872
GO:0005524
GO:0016849
GO:0009190
GO:0004930
GO:0007166
GO:0003723
GO:0071315
GO:0098685
GO:0007626
GO:0016323
GO:0051480
GO:0010255
GO:1900273
GO:1900454
GO:0008294
GO:0035774
GO:0098686
GO:0032793
GO:0031915
GO:0060076
GO:0150076
GO:0016324
GO:0014069
GO:0048786
GO:0030425
GO:0030424
GO:0080135
GO:0007616
GO:0098978
GO:0032809
GO:0099056
GO:0005887
GO:0007189
GO:0016199
GO:0031290
GO:0016829
GO:0016020
GO:0016876
GO:0043039
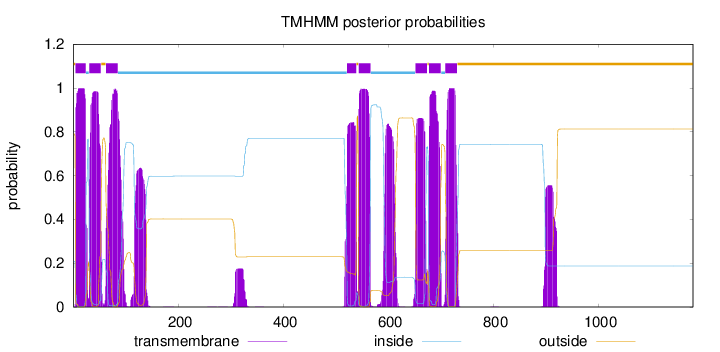
Topology
Length:
1180
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
215.365600000001
Exp number, first 60 AAs:
42.27683
Total prob of N-in:
0.20947
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 24
inside
25 - 30
TMhelix
31 - 53
outside
54 - 62
TMhelix
63 - 85
inside
86 - 521
TMhelix
522 - 539
outside
540 - 543
TMhelix
544 - 566
inside
567 - 651
TMhelix
652 - 674
outside
675 - 677
TMhelix
678 - 700
inside
701 - 708
TMhelix
709 - 731
outside
732 - 1180
Population Genetic Test Statistics
Pi
241.949484
Theta
173.567221
Tajima's D
1.974035
CLR
0.314374
CSRT
0.875206239688016
Interpretation
Uncertain
