Gene
KWMTBOMO09275
Pre Gene Modal
BGIBMGA003422
Annotation
hypothetical_protein_OBRU01_13126_[Operophtera_brumata]
Full name
Gamma-tubulin complex component
Location in the cell
Mitochondrial Reliability : 1.686
Sequence
CDS
ATGGCTGCGGCTTCGAGAAAATCGCTTGGACAGCTAATTCAACAGGGATGGAACGAGATCCCTGAAATTTTTGCCTCAACAGGCTTAGGTCTTGTTGGTATTGGTCTGGGTGTTCTTGGAGCATACAATTACGTGAAGAACGATGGAGACAATAGAAGGTACAAGGGTACCTATGTGGTGATGAGGCCGGACGACCCAAGAGTGAAGCTTATACGCAAGGAGTAA
Protein
MAAASRKSLGQLIQQGWNEIPEIFASTGLGLVGIGLGVLGAYNYVKNDGDNRRYKGTYVVMRPDDPRVKLIRKE
Summary
Similarity
Belongs to the TUBGCP family.
Uniprot
H9J1N5
A0A2H1VKS4
A0A2W1BAZ9
A0A0L7L8V3
S4NVN0
A0A194PR41
+ More
A0A194RJ06 A0A2M3Z3X9 A0A182QZ54 A0A182J4H4 A0A182FSV4 A0A182I8M0 D3TS47 T1DF07 A0A2M4AFJ0 A0A2M4C5R2 A0A084W782 A0A182XX58 A0A182P2Q0 A0A182NG01 A0A182WUZ5 Q7PS85 A0A182RRT8 A0A182VIH5 A0A1A9UHV9 A0A182KBQ8 W5J5J8 A0A1Q3FKH9 A0A1B0BMN8 A0A194PLI6 A0A194RJS8 B0W6P0 A0A1A9ZB87 A0A1L8EER6 A0A1I8P6Q2 A0A067R1G1 A0A023ED60 Q1HRA0 A0A1I8MHI4 B4PZW7 B3NUG7 A0A1W4UQJ5 B3MZP9 A0A1A9WB06 A0A0L7L8F7 U5EP65 Q29GX9 B4H2F0 B4M3R2 B4JXL9 B4L385 B4NEU5 A0A3B0K2P6 A0A1L8DUY8 B4R6U7 B4IJS0 Q9W380 A0A1B0D6S6 A0A0M3QZG1 A0A139WIC3 A0A1W4XVM0 A0A1D2MKI9 A0A232EM37 T1GX67 A0A336MFN9 A0A182W138 A0A2S2QK80
A0A194RJ06 A0A2M3Z3X9 A0A182QZ54 A0A182J4H4 A0A182FSV4 A0A182I8M0 D3TS47 T1DF07 A0A2M4AFJ0 A0A2M4C5R2 A0A084W782 A0A182XX58 A0A182P2Q0 A0A182NG01 A0A182WUZ5 Q7PS85 A0A182RRT8 A0A182VIH5 A0A1A9UHV9 A0A182KBQ8 W5J5J8 A0A1Q3FKH9 A0A1B0BMN8 A0A194PLI6 A0A194RJS8 B0W6P0 A0A1A9ZB87 A0A1L8EER6 A0A1I8P6Q2 A0A067R1G1 A0A023ED60 Q1HRA0 A0A1I8MHI4 B4PZW7 B3NUG7 A0A1W4UQJ5 B3MZP9 A0A1A9WB06 A0A0L7L8F7 U5EP65 Q29GX9 B4H2F0 B4M3R2 B4JXL9 B4L385 B4NEU5 A0A3B0K2P6 A0A1L8DUY8 B4R6U7 B4IJS0 Q9W380 A0A1B0D6S6 A0A0M3QZG1 A0A139WIC3 A0A1W4XVM0 A0A1D2MKI9 A0A232EM37 T1GX67 A0A336MFN9 A0A182W138 A0A2S2QK80
Pubmed
19121390
28756777
26227816
23622113
26354079
20353571
+ More
24438588 25244985 12364791 14747013 17210077 20920257 23761445 24845553 24945155 26483478 17204158 17510324 25315136 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 27289101 28648823
24438588 25244985 12364791 14747013 17210077 20920257 23761445 24845553 24945155 26483478 17204158 17510324 25315136 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 27289101 28648823
EMBL
BABH01023936
ODYU01002854
ODYU01003096
SOQ40854.1
SOQ41418.1
KZ150539
+ More
PZC70577.1 JTDY01002253 KOB71790.1 GAIX01011476 JAA81084.1 KQ459602 KPI93595.1 KQ460106 KPJ17808.1 GGFM01002452 MBW23203.1 AXCN02000343 APCN01003086 CCAG010009137 EZ424249 ADD20525.1 GAMD01003301 JAA98289.1 GGFK01006161 MBW39482.1 GGFJ01011350 MBW60491.1 ATLV01021173 KE525314 KFB46076.1 AAAB01008844 EAA06017.4 ADMH02002125 ETN58673.1 GFDL01006934 JAV28111.1 JXJN01017007 KPI93594.1 KPJ17807.1 DS231849 EDS36871.1 GFDG01001587 JAV17212.1 KK852815 KDR15806.1 JXUM01104763 GAPW01006788 KQ564912 JAC06810.1 KXJ71583.1 DQ440194 CH477193 ABF18227.1 EAT48534.1 EAT48535.1 CM000162 EDX02173.1 CH954180 EDV46290.1 CH902635 EDV33850.1 KOB71787.1 GANO01004864 JAB55007.1 CH379064 EAL31980.1 CH479204 EDW30517.1 CH940651 EDW65437.2 CH916376 EDV95118.1 CH933810 EDW07013.1 CH964239 EDW82264.1 OUUW01000015 SPP88537.1 GFDF01003830 JAV10254.1 CM000366 EDX17477.1 CH480849 EDW51278.1 AE014298 AY071733 AAF46455.1 AAL49355.1 AJVK01026235 CP012528 ALC49363.1 KQ971342 KYB27682.1 LJIJ01000956 ODM93549.1 NNAY01003442 OXU19407.1 CAQQ02091942 UFQS01001166 UFQT01001166 SSX09320.1 SSX29222.1 GGMS01008389 MBY77592.1
PZC70577.1 JTDY01002253 KOB71790.1 GAIX01011476 JAA81084.1 KQ459602 KPI93595.1 KQ460106 KPJ17808.1 GGFM01002452 MBW23203.1 AXCN02000343 APCN01003086 CCAG010009137 EZ424249 ADD20525.1 GAMD01003301 JAA98289.1 GGFK01006161 MBW39482.1 GGFJ01011350 MBW60491.1 ATLV01021173 KE525314 KFB46076.1 AAAB01008844 EAA06017.4 ADMH02002125 ETN58673.1 GFDL01006934 JAV28111.1 JXJN01017007 KPI93594.1 KPJ17807.1 DS231849 EDS36871.1 GFDG01001587 JAV17212.1 KK852815 KDR15806.1 JXUM01104763 GAPW01006788 KQ564912 JAC06810.1 KXJ71583.1 DQ440194 CH477193 ABF18227.1 EAT48534.1 EAT48535.1 CM000162 EDX02173.1 CH954180 EDV46290.1 CH902635 EDV33850.1 KOB71787.1 GANO01004864 JAB55007.1 CH379064 EAL31980.1 CH479204 EDW30517.1 CH940651 EDW65437.2 CH916376 EDV95118.1 CH933810 EDW07013.1 CH964239 EDW82264.1 OUUW01000015 SPP88537.1 GFDF01003830 JAV10254.1 CM000366 EDX17477.1 CH480849 EDW51278.1 AE014298 AY071733 AAF46455.1 AAL49355.1 AJVK01026235 CP012528 ALC49363.1 KQ971342 KYB27682.1 LJIJ01000956 ODM93549.1 NNAY01003442 OXU19407.1 CAQQ02091942 UFQS01001166 UFQT01001166 SSX09320.1 SSX29222.1 GGMS01008389 MBY77592.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000075886
UP000075880
+ More
UP000069272 UP000075840 UP000092444 UP000030765 UP000076408 UP000075885 UP000075884 UP000076407 UP000007062 UP000075900 UP000075903 UP000078200 UP000075881 UP000000673 UP000092460 UP000002320 UP000092445 UP000095300 UP000027135 UP000069940 UP000249989 UP000008820 UP000095301 UP000002282 UP000008711 UP000192221 UP000007801 UP000091820 UP000001819 UP000008744 UP000008792 UP000001070 UP000009192 UP000007798 UP000268350 UP000000304 UP000001292 UP000000803 UP000092462 UP000092553 UP000007266 UP000192223 UP000094527 UP000215335 UP000015102 UP000075920
UP000069272 UP000075840 UP000092444 UP000030765 UP000076408 UP000075885 UP000075884 UP000076407 UP000007062 UP000075900 UP000075903 UP000078200 UP000075881 UP000000673 UP000092460 UP000002320 UP000092445 UP000095300 UP000027135 UP000069940 UP000249989 UP000008820 UP000095301 UP000002282 UP000008711 UP000192221 UP000007801 UP000091820 UP000001819 UP000008744 UP000008792 UP000001070 UP000009192 UP000007798 UP000268350 UP000000304 UP000001292 UP000000803 UP000092462 UP000092553 UP000007266 UP000192223 UP000094527 UP000215335 UP000015102 UP000075920
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9J1N5
A0A2H1VKS4
A0A2W1BAZ9
A0A0L7L8V3
S4NVN0
A0A194PR41
+ More
A0A194RJ06 A0A2M3Z3X9 A0A182QZ54 A0A182J4H4 A0A182FSV4 A0A182I8M0 D3TS47 T1DF07 A0A2M4AFJ0 A0A2M4C5R2 A0A084W782 A0A182XX58 A0A182P2Q0 A0A182NG01 A0A182WUZ5 Q7PS85 A0A182RRT8 A0A182VIH5 A0A1A9UHV9 A0A182KBQ8 W5J5J8 A0A1Q3FKH9 A0A1B0BMN8 A0A194PLI6 A0A194RJS8 B0W6P0 A0A1A9ZB87 A0A1L8EER6 A0A1I8P6Q2 A0A067R1G1 A0A023ED60 Q1HRA0 A0A1I8MHI4 B4PZW7 B3NUG7 A0A1W4UQJ5 B3MZP9 A0A1A9WB06 A0A0L7L8F7 U5EP65 Q29GX9 B4H2F0 B4M3R2 B4JXL9 B4L385 B4NEU5 A0A3B0K2P6 A0A1L8DUY8 B4R6U7 B4IJS0 Q9W380 A0A1B0D6S6 A0A0M3QZG1 A0A139WIC3 A0A1W4XVM0 A0A1D2MKI9 A0A232EM37 T1GX67 A0A336MFN9 A0A182W138 A0A2S2QK80
A0A194RJ06 A0A2M3Z3X9 A0A182QZ54 A0A182J4H4 A0A182FSV4 A0A182I8M0 D3TS47 T1DF07 A0A2M4AFJ0 A0A2M4C5R2 A0A084W782 A0A182XX58 A0A182P2Q0 A0A182NG01 A0A182WUZ5 Q7PS85 A0A182RRT8 A0A182VIH5 A0A1A9UHV9 A0A182KBQ8 W5J5J8 A0A1Q3FKH9 A0A1B0BMN8 A0A194PLI6 A0A194RJS8 B0W6P0 A0A1A9ZB87 A0A1L8EER6 A0A1I8P6Q2 A0A067R1G1 A0A023ED60 Q1HRA0 A0A1I8MHI4 B4PZW7 B3NUG7 A0A1W4UQJ5 B3MZP9 A0A1A9WB06 A0A0L7L8F7 U5EP65 Q29GX9 B4H2F0 B4M3R2 B4JXL9 B4L385 B4NEU5 A0A3B0K2P6 A0A1L8DUY8 B4R6U7 B4IJS0 Q9W380 A0A1B0D6S6 A0A0M3QZG1 A0A139WIC3 A0A1W4XVM0 A0A1D2MKI9 A0A232EM37 T1GX67 A0A336MFN9 A0A182W138 A0A2S2QK80
Ontologies
GO
PANTHER
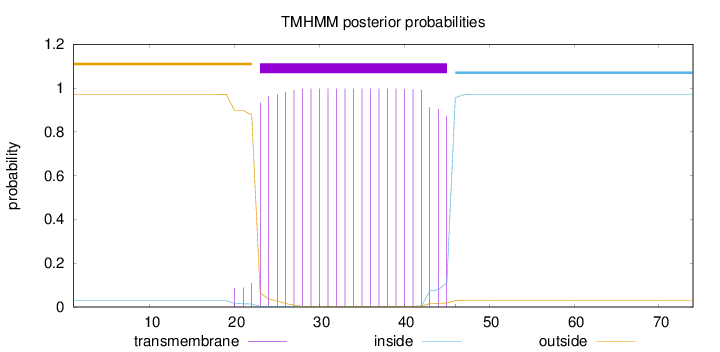
Topology
Subcellular location
Length:
74
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.77804
Exp number, first 60 AAs:
22.77804
Total prob of N-in:
0.02875
POSSIBLE N-term signal
sequence
outside
1 - 22
TMhelix
23 - 45
inside
46 - 74
Population Genetic Test Statistics
Pi
239.614347
Theta
174.742014
Tajima's D
1.167895
CLR
0
CSRT
0.709314534273286
Interpretation
Uncertain
