Gene
KWMTBOMO09274 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003319
Annotation
PREDICTED:_aspartate_aminotransferase?_cytoplasmic_isoform_X1_[Amyelois_transitella]
Full name
Aspartate aminotransferase
Location in the cell
Cytoplasmic Reliability : 2.056
Sequence
CDS
ATGGCTTCTCGATTCCAAGGTGTCGAACAAGGCCCCCCGATAGAGGTGTTTTTGTTAAACAGACTTTTTACTGAGGATACTTTTCAAAATAAAGTTAATTTAGGAGTTGGAGCATACAGAGATGAAAATGGAAAACCATGGGTACTGCCCATCGTTCGTAAGATGGAAAAACAGCTGGCAGCAGATGAAACCTTACTGCATGAATACCTTCCCGTGTTAGGTCTGGAGCAGTTTTGTAATGCGTCTGTGGCTATGCTCCTTGGTGAGGATAGTCCTGCCATCGCCGCTGGAAAAGCTTTCGGTGTGCAAGTCTTGTCCGGGACTGGTGGTCTGCGCGTCGGCGCCGAGTTCCTCAACAAGCATCTTAAATATGATACATTCTACTACTCCACACCTACATGGGAGAATCATCACCTAGTATTCGTGAACTCTGGTTTCACGAAGCCTCGCTCGTACCGCTACTGGGACCCGAAGACCAGGGGCATAGACTTCGACGGTTTCATCGAAGACCTCAAGAGCGCTCCGGAGAATGCCGTCATACTGCTGCACGCATGCGCCCACAATCCCACAGGCATAGACCCCACCAGGGAACAGTGGATGAAAATAGCTGATGTTATGGAGGAGCGCAAGTTGTTCCCGTTCTTCGACAGCGCGTACCAAGGGTTCGCGTCGGGCGACTTGGACGGAGACGCGTGGGCGGTGCGGTACTTCGTCAGTCGCGGCTTCGAACTCGTCTGCGCTCAGTCCTATGCCAAGAATTTCGGACTTTACAATGAACGCGTTGGCAACCTAGCGGTGGTATTGAACGAGTCTGCGCACGTTGCTCGCATAAAGTCCCAGATGACGTGGATCGTGCGAGGGATGTACTCCAACCCGCCAGCGCACGGGGCCCACGTCGTGGCCGCCGTGCTCACTAACAAGCAGCTCTTCGACGAGTGGAGGGACCACATCAAACTCATGTCTACGAGGGTCATGCAGATGAGAGAAGCTCTGAGGTCTGAGCTCGTGAAACTGGGAACTCCCGGCAACTGGGACCACATAGTGAAGCAGATTGGGCTATTCTCGTACACGGGTCTGAACCAGCGCCAGTGCGAGTACTTGATACAGGAGTACCACATCTATCTGCTGAAGTCCGGTCGCATCAACATCTGCGGGCTGAACCCGGGAAACGTGCAGTACGTGGCTAAAGCCATCAACGACGCGGTCACCAAGTTCCCTGACGATCACGACAATCGAGCGAAGTTATGA
Protein
MASRFQGVEQGPPIEVFLLNRLFTEDTFQNKVNLGVGAYRDENGKPWVLPIVRKMEKQLAADETLLHEYLPVLGLEQFCNASVAMLLGEDSPAIAAGKAFGVQVLSGTGGLRVGAEFLNKHLKYDTFYYSTPTWENHHLVFVNSGFTKPRSYRYWDPKTRGIDFDGFIEDLKSAPENAVILLHACAHNPTGIDPTREQWMKIADVMEERKLFPFFDSAYQGFASGDLDGDAWAVRYFVSRGFELVCAQSYAKNFGLYNERVGNLAVVLNESAHVARIKSQMTWIVRGMYSNPPAHGAHVVAAVLTNKQLFDEWRDHIKLMSTRVMQMREALRSELVKLGTPGNWDHIVKQIGLFSYTGLNQRQCEYLIQEYHIYLLKSGRINICGLNPGNVQYVAKAINDAVTKFPDDHDNRAKL
Summary
Catalytic Activity
2-oxoglutarate + L-aspartate = L-glutamate + oxaloacetate
Subunit
Homodimer.
Miscellaneous
In eukaryotes there are cytoplasmic, mitochondrial and chloroplastic isozymes.
Feature
chain Aspartate aminotransferase
Uniprot
H9J1D2
A0A212ENG7
A0A2A4JUG9
A0A194PKR4
A0A2A4JUS7
S4P8E3
+ More
A0A2W1B2Y8 A0A437BNU4 A0A286T886 A0A1B6LUX2 E2AD28 A0A232F6X6 K7J4Q5 A0A1I8MDM9 A0A0L7QQR4 A0A1L8EHE6 J3JZG8 A0A336ME91 A0A139WGX0 D6WLK0 A0A026VVN6 A0A1I8NTS8 F4WRA2 A0A1I8NTU3 B0WKE5 G3MNQ8 A0A423U3M3 A0A0J7NT78 A0A0L0CPL8 A0A1B6EGT9 E0VIU2 A0A1Y1LB57 E2BMX0 A0A0T6BEY3 A0A411G7V3 A0A154PH48 V5SI13 A0A023FW59 A0A087UAJ9 A0A1A9W3A5 A0A1Q3FJP1 Q5K6H4 A0A023ERN7 A0A023ESU6 A0A158NUA6 A0A182J070 A0A2M4AD35 A0A286T8E3 A0A182F9E4 A0A084VLB6 A0A1B0B6M6 A0A131XIE3 A0A182RYK7 A0A182MB16 Q16LN3 A0A182PPZ4 A0A1B0A700 A0A2M4BQ28 A0A1E1XHD6 W5JI36 A0A1L8E2F8 A0A182N5Q7 A0A1A9UIE9 A0A1L8DQY1 D3TRZ7 A0A182Q0R3 A0A3B0J1W3 A0A1D2MYX3 A0A182W1K6 A0A182U2L9 A0A182YGL0 A0A182UP30 A0A182L7F0 Q7QB79 L7M7N7 C1C1U3 A0A182XJ05 A0A067RE69 A0A182I7V0 A0A0K8RKI9 U5EW46 A0A2A3EF32 V9IKH9 A0A2R5L684 A0A293MQX8 A0A0P5K381 A0A0P5NY43 A0A0P5HRT9 A0A0P6AZD2 A0A0P5I1C1 A0A0P5J4N6 B5E189 B4H584 A0A0P5IAY4 A0A0P5RQV0 E9H8T5 V5G1F1 A0A0P5HV36 A0A0P6FG42 A0A0P5K316
A0A2W1B2Y8 A0A437BNU4 A0A286T886 A0A1B6LUX2 E2AD28 A0A232F6X6 K7J4Q5 A0A1I8MDM9 A0A0L7QQR4 A0A1L8EHE6 J3JZG8 A0A336ME91 A0A139WGX0 D6WLK0 A0A026VVN6 A0A1I8NTS8 F4WRA2 A0A1I8NTU3 B0WKE5 G3MNQ8 A0A423U3M3 A0A0J7NT78 A0A0L0CPL8 A0A1B6EGT9 E0VIU2 A0A1Y1LB57 E2BMX0 A0A0T6BEY3 A0A411G7V3 A0A154PH48 V5SI13 A0A023FW59 A0A087UAJ9 A0A1A9W3A5 A0A1Q3FJP1 Q5K6H4 A0A023ERN7 A0A023ESU6 A0A158NUA6 A0A182J070 A0A2M4AD35 A0A286T8E3 A0A182F9E4 A0A084VLB6 A0A1B0B6M6 A0A131XIE3 A0A182RYK7 A0A182MB16 Q16LN3 A0A182PPZ4 A0A1B0A700 A0A2M4BQ28 A0A1E1XHD6 W5JI36 A0A1L8E2F8 A0A182N5Q7 A0A1A9UIE9 A0A1L8DQY1 D3TRZ7 A0A182Q0R3 A0A3B0J1W3 A0A1D2MYX3 A0A182W1K6 A0A182U2L9 A0A182YGL0 A0A182UP30 A0A182L7F0 Q7QB79 L7M7N7 C1C1U3 A0A182XJ05 A0A067RE69 A0A182I7V0 A0A0K8RKI9 U5EW46 A0A2A3EF32 V9IKH9 A0A2R5L684 A0A293MQX8 A0A0P5K381 A0A0P5NY43 A0A0P5HRT9 A0A0P6AZD2 A0A0P5I1C1 A0A0P5J4N6 B5E189 B4H584 A0A0P5IAY4 A0A0P5RQV0 E9H8T5 V5G1F1 A0A0P5HV36 A0A0P6FG42 A0A0P5K316
EC Number
2.6.1.1
Pubmed
19121390
22118469
26354079
23622113
28756777
20798317
+ More
28648823 20075255 25315136 22516182 23537049 18362917 19820115 24508170 30249741 21719571 22216098 26108605 20566863 28004739 24945155 21347285 24438588 28049606 17510324 28503490 20920257 23761445 20353571 27289101 25244985 20966253 12364791 14747013 17210077 25576852 24845553 15632085 17994087 21292972
28648823 20075255 25315136 22516182 23537049 18362917 19820115 24508170 30249741 21719571 22216098 26108605 20566863 28004739 24945155 21347285 24438588 28049606 17510324 28503490 20920257 23761445 20353571 27289101 25244985 20966253 12364791 14747013 17210077 25576852 24845553 15632085 17994087 21292972
EMBL
BABH01023934
AGBW02013658
OWR43034.1
NWSH01000596
PCG75449.1
KQ459602
+ More
KPI93593.1 PCG75448.1 GAIX01004219 JAA88341.1 KZ150539 PZC70578.1 RSAL01000026 RVE52055.1 LC260175 BBA45746.1 GEBQ01012506 GEBQ01004393 JAT27471.1 JAT35584.1 GL438658 EFN68664.1 NNAY01000855 OXU26188.1 AAZX01008388 KQ414785 KOC60967.1 GFDG01000639 JAV18160.1 APGK01028345 BT128650 KB740686 KB631930 AEE63607.1 ENN79554.1 ERL87311.1 UFQT01000773 SSX27113.1 KQ971343 KYB27212.1 EFA04122.1 KK107796 QOIP01000013 EZA47715.1 RLU15567.1 GL888284 EGI63319.1 DS231971 EDS29765.1 JO843509 AEO35126.1 QCYY01000703 ROT83300.1 LBMM01001886 KMQ95610.1 JRES01000080 KNC34305.1 GEDC01000146 JAS37152.1 DS235206 EEB13298.1 GEZM01063258 JAV69165.1 GL449377 EFN82952.1 LJIG01001042 KRT85898.1 MH366205 QBB02014.1 KQ434902 KZC11179.1 KF647648 AHB50510.1 GBBL01001234 JAC26086.1 KK119009 KFM74388.1 GFDL01007248 JAV27797.1 AF395205 AAQ02891.1 GAPW01001728 JAC11870.1 GAPW01001727 JAC11871.1 ADTU01026372 GGFK01005372 MBW38693.1 LC260176 BBA45747.1 ATLV01014443 KE524972 KFB38760.1 JXJN01009163 GEFH01001668 JAP66913.1 AXCM01000329 CH477898 EAT35255.1 GGFJ01005973 MBW55114.1 GFAC01000702 JAT98486.1 ADMH02001121 ETN64052.1 GFDF01001275 JAV12809.1 GFDF01005273 JAV08811.1 EZ424199 ADD20475.1 AXCN02001158 OUUW01000001 SPP74995.1 LJIJ01000374 ODM98200.1 AAAB01008880 EAA08515.4 GACK01004718 JAA60316.1 BT080822 ACO15246.1 KK852561 KDR21333.1 APCN01002556 GADI01002739 JAA71069.1 GANO01001584 JAB58287.1 KZ288271 PBC29766.1 JR048984 AEY60824.1 GGLE01000821 MBY04947.1 GFWV01018565 MAA43293.1 GDIQ01190268 JAK61457.1 GDIQ01140540 JAL11186.1 GDIQ01225852 JAK25873.1 GDIP01022172 JAM81543.1 GDIQ01222057 JAK29668.1 GDIQ01206664 JAK45061.1 CM000071 EDY69477.1 CH479210 EDW32920.1 GDIQ01216008 JAK35717.1 GDIQ01098149 JAL53577.1 GL732605 EFX71867.1 GALX01004621 JAB63845.1 GDIQ01224700 JAK27025.1 GDIQ01047870 JAN46867.1 GDIQ01216007 JAK35718.1
KPI93593.1 PCG75448.1 GAIX01004219 JAA88341.1 KZ150539 PZC70578.1 RSAL01000026 RVE52055.1 LC260175 BBA45746.1 GEBQ01012506 GEBQ01004393 JAT27471.1 JAT35584.1 GL438658 EFN68664.1 NNAY01000855 OXU26188.1 AAZX01008388 KQ414785 KOC60967.1 GFDG01000639 JAV18160.1 APGK01028345 BT128650 KB740686 KB631930 AEE63607.1 ENN79554.1 ERL87311.1 UFQT01000773 SSX27113.1 KQ971343 KYB27212.1 EFA04122.1 KK107796 QOIP01000013 EZA47715.1 RLU15567.1 GL888284 EGI63319.1 DS231971 EDS29765.1 JO843509 AEO35126.1 QCYY01000703 ROT83300.1 LBMM01001886 KMQ95610.1 JRES01000080 KNC34305.1 GEDC01000146 JAS37152.1 DS235206 EEB13298.1 GEZM01063258 JAV69165.1 GL449377 EFN82952.1 LJIG01001042 KRT85898.1 MH366205 QBB02014.1 KQ434902 KZC11179.1 KF647648 AHB50510.1 GBBL01001234 JAC26086.1 KK119009 KFM74388.1 GFDL01007248 JAV27797.1 AF395205 AAQ02891.1 GAPW01001728 JAC11870.1 GAPW01001727 JAC11871.1 ADTU01026372 GGFK01005372 MBW38693.1 LC260176 BBA45747.1 ATLV01014443 KE524972 KFB38760.1 JXJN01009163 GEFH01001668 JAP66913.1 AXCM01000329 CH477898 EAT35255.1 GGFJ01005973 MBW55114.1 GFAC01000702 JAT98486.1 ADMH02001121 ETN64052.1 GFDF01001275 JAV12809.1 GFDF01005273 JAV08811.1 EZ424199 ADD20475.1 AXCN02001158 OUUW01000001 SPP74995.1 LJIJ01000374 ODM98200.1 AAAB01008880 EAA08515.4 GACK01004718 JAA60316.1 BT080822 ACO15246.1 KK852561 KDR21333.1 APCN01002556 GADI01002739 JAA71069.1 GANO01001584 JAB58287.1 KZ288271 PBC29766.1 JR048984 AEY60824.1 GGLE01000821 MBY04947.1 GFWV01018565 MAA43293.1 GDIQ01190268 JAK61457.1 GDIQ01140540 JAL11186.1 GDIQ01225852 JAK25873.1 GDIP01022172 JAM81543.1 GDIQ01222057 JAK29668.1 GDIQ01206664 JAK45061.1 CM000071 EDY69477.1 CH479210 EDW32920.1 GDIQ01216008 JAK35717.1 GDIQ01098149 JAL53577.1 GL732605 EFX71867.1 GALX01004621 JAB63845.1 GDIQ01224700 JAK27025.1 GDIQ01047870 JAN46867.1 GDIQ01216007 JAK35718.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000283053
UP000000311
+ More
UP000215335 UP000002358 UP000095301 UP000053825 UP000019118 UP000030742 UP000007266 UP000053097 UP000279307 UP000095300 UP000007755 UP000002320 UP000283509 UP000036403 UP000037069 UP000009046 UP000008237 UP000076502 UP000054359 UP000091820 UP000005205 UP000075880 UP000069272 UP000030765 UP000092460 UP000075900 UP000075883 UP000008820 UP000075885 UP000092445 UP000000673 UP000075884 UP000078200 UP000075886 UP000268350 UP000094527 UP000075920 UP000075902 UP000076408 UP000075903 UP000075882 UP000007062 UP000076407 UP000027135 UP000075840 UP000242457 UP000001819 UP000008744 UP000000305
UP000215335 UP000002358 UP000095301 UP000053825 UP000019118 UP000030742 UP000007266 UP000053097 UP000279307 UP000095300 UP000007755 UP000002320 UP000283509 UP000036403 UP000037069 UP000009046 UP000008237 UP000076502 UP000054359 UP000091820 UP000005205 UP000075880 UP000069272 UP000030765 UP000092460 UP000075900 UP000075883 UP000008820 UP000075885 UP000092445 UP000000673 UP000075884 UP000078200 UP000075886 UP000268350 UP000094527 UP000075920 UP000075902 UP000076408 UP000075903 UP000075882 UP000007062 UP000076407 UP000027135 UP000075840 UP000242457 UP000001819 UP000008744 UP000000305
Pfam
PF00155 Aminotran_1_2
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9J1D2
A0A212ENG7
A0A2A4JUG9
A0A194PKR4
A0A2A4JUS7
S4P8E3
+ More
A0A2W1B2Y8 A0A437BNU4 A0A286T886 A0A1B6LUX2 E2AD28 A0A232F6X6 K7J4Q5 A0A1I8MDM9 A0A0L7QQR4 A0A1L8EHE6 J3JZG8 A0A336ME91 A0A139WGX0 D6WLK0 A0A026VVN6 A0A1I8NTS8 F4WRA2 A0A1I8NTU3 B0WKE5 G3MNQ8 A0A423U3M3 A0A0J7NT78 A0A0L0CPL8 A0A1B6EGT9 E0VIU2 A0A1Y1LB57 E2BMX0 A0A0T6BEY3 A0A411G7V3 A0A154PH48 V5SI13 A0A023FW59 A0A087UAJ9 A0A1A9W3A5 A0A1Q3FJP1 Q5K6H4 A0A023ERN7 A0A023ESU6 A0A158NUA6 A0A182J070 A0A2M4AD35 A0A286T8E3 A0A182F9E4 A0A084VLB6 A0A1B0B6M6 A0A131XIE3 A0A182RYK7 A0A182MB16 Q16LN3 A0A182PPZ4 A0A1B0A700 A0A2M4BQ28 A0A1E1XHD6 W5JI36 A0A1L8E2F8 A0A182N5Q7 A0A1A9UIE9 A0A1L8DQY1 D3TRZ7 A0A182Q0R3 A0A3B0J1W3 A0A1D2MYX3 A0A182W1K6 A0A182U2L9 A0A182YGL0 A0A182UP30 A0A182L7F0 Q7QB79 L7M7N7 C1C1U3 A0A182XJ05 A0A067RE69 A0A182I7V0 A0A0K8RKI9 U5EW46 A0A2A3EF32 V9IKH9 A0A2R5L684 A0A293MQX8 A0A0P5K381 A0A0P5NY43 A0A0P5HRT9 A0A0P6AZD2 A0A0P5I1C1 A0A0P5J4N6 B5E189 B4H584 A0A0P5IAY4 A0A0P5RQV0 E9H8T5 V5G1F1 A0A0P5HV36 A0A0P6FG42 A0A0P5K316
A0A2W1B2Y8 A0A437BNU4 A0A286T886 A0A1B6LUX2 E2AD28 A0A232F6X6 K7J4Q5 A0A1I8MDM9 A0A0L7QQR4 A0A1L8EHE6 J3JZG8 A0A336ME91 A0A139WGX0 D6WLK0 A0A026VVN6 A0A1I8NTS8 F4WRA2 A0A1I8NTU3 B0WKE5 G3MNQ8 A0A423U3M3 A0A0J7NT78 A0A0L0CPL8 A0A1B6EGT9 E0VIU2 A0A1Y1LB57 E2BMX0 A0A0T6BEY3 A0A411G7V3 A0A154PH48 V5SI13 A0A023FW59 A0A087UAJ9 A0A1A9W3A5 A0A1Q3FJP1 Q5K6H4 A0A023ERN7 A0A023ESU6 A0A158NUA6 A0A182J070 A0A2M4AD35 A0A286T8E3 A0A182F9E4 A0A084VLB6 A0A1B0B6M6 A0A131XIE3 A0A182RYK7 A0A182MB16 Q16LN3 A0A182PPZ4 A0A1B0A700 A0A2M4BQ28 A0A1E1XHD6 W5JI36 A0A1L8E2F8 A0A182N5Q7 A0A1A9UIE9 A0A1L8DQY1 D3TRZ7 A0A182Q0R3 A0A3B0J1W3 A0A1D2MYX3 A0A182W1K6 A0A182U2L9 A0A182YGL0 A0A182UP30 A0A182L7F0 Q7QB79 L7M7N7 C1C1U3 A0A182XJ05 A0A067RE69 A0A182I7V0 A0A0K8RKI9 U5EW46 A0A2A3EF32 V9IKH9 A0A2R5L684 A0A293MQX8 A0A0P5K381 A0A0P5NY43 A0A0P5HRT9 A0A0P6AZD2 A0A0P5I1C1 A0A0P5J4N6 B5E189 B4H584 A0A0P5IAY4 A0A0P5RQV0 E9H8T5 V5G1F1 A0A0P5HV36 A0A0P6FG42 A0A0P5K316
PDB
2CST
E-value=1.93691e-140,
Score=1279
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00400 Phenylalanine, tyrosine and tryptophan biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00330 Arginine and proline metabolism - Bombyx mori (domestic silkworm)
00350 Tyrosine metabolism - Bombyx mori (domestic silkworm)
00360 Phenylalanine metabolism - Bombyx mori (domestic silkworm)
00400 Phenylalanine, tyrosine and tryptophan biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
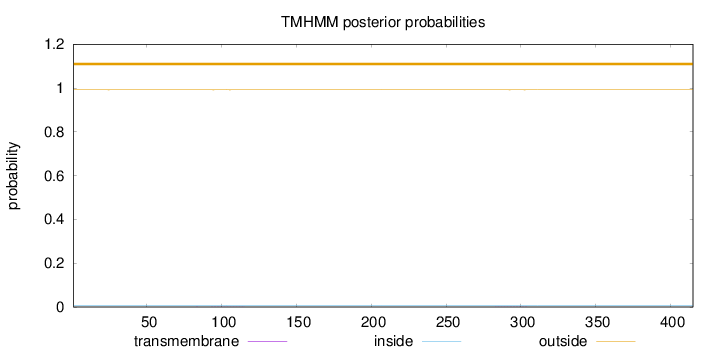
Topology
Length:
415
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01437
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00814
outside
1 - 415
Population Genetic Test Statistics
Pi
221.199277
Theta
169.517682
Tajima's D
0.98518
CLR
0.528907
CSRT
0.654367281635918
Interpretation
Uncertain
