Gene
KWMTBOMO09250 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003330
Annotation
putative_inosine-uridine_preferring_nucleoside_hydrolase_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 2.922
Sequence
CDS
ATGTATTCGAGAATATTCGTGTATTTGGTTAAGTTATTTTTTTTTCTCAGCTGTAAATCGGAAGCTCGAATTAATGACGTCACCGAAGCCAGAATCCTCATTGACCACGACGGAGGTGCCGATGATTCTTTAGCGATATTCACAGCGCTGCTGTATGAGGAGAAATTCAATGGACCGAAAGTCATAGGTTTGACTACAACACACGGTAACGTCAATGAAGATCAAGTTTACGTAAACAGTCAAAGAATATTGGGTGTGGCGAACAGAAGAGATGTACCGATCTACAGAGGCTGCTCTGAGCCACTAATTGGAGGTTCTGTTAGTGACTTCTACTTTGGAAACGATGGTCTGGGAGATCATGAGAACGTTGAATACCAGAGCATTGCGGCTGAGAAGGAGCACGCCGTTTTTCGTATAATTGAACTGTCAAAATTATATCCAGGTAATTTGGAAATAATCGCTATTGGTCCCTTGACTAATATTGCTATGGCTGTGAAAATTGATCCGTTTTTCCTAAGTCGACTATCACGCCTAGTTATAGGTGCTGGGTATATATACAGTGATGACTACAAAGAACCAGAGTTCAATGTGGATATGGATGTCGAAGCATACCACATCGTGACAAGAAGCGCAAGCCCTGATATAGTCACCATACTGCCTTTCTCGCAAGTCCGAACATGGTTGAATATTAGTAATGAATGGCGCAAGCAAACTCTGGGTTCAATTAATACGGATATAATGAAAGCTCAGAATTCTTACGAACGAAAATCATTGAGTAAAAGCCGTTGTTGGTCGACCCTAGACCCTGCCGTTGTTGCCATAGCTCTGGACGAGACGATCGCTGAGGAGACTGTCTTCTCAAACAACAGTGTTGTACTATGCGGTTCAAACCGCGGCGTTGTCTTGAATGATTTTGGCTCAGAACCAAACGTCAAACTGTGGTACAGACTTAACATGCAAAAGTACAAGCAATTCTTGTTGCGAGTGTTTAGTGTTGAATAA
Protein
MYSRIFVYLVKLFFFLSCKSEARINDVTEARILIDHDGGADDSLAIFTALLYEEKFNGPKVIGLTTTHGNVNEDQVYVNSQRILGVANRRDVPIYRGCSEPLIGGSVSDFYFGNDGLGDHENVEYQSIAAEKEHAVFRIIELSKLYPGNLEIIAIGPLTNIAMAVKIDPFFLSRLSRLVIGAGYIYSDDYKEPEFNVDMDVEAYHIVTRSASPDIVTILPFSQVRTWLNISNEWRKQTLGSINTDIMKAQNSYERKSLSKSRCWSTLDPAVVAIALDETIAEETVFSNNSVVLCGSNRGVVLNDFGSEPNVKLWYRLNMQKYKQFLLRVFSVE
Summary
Similarity
Belongs to the CRISP family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the IUNH family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the IUNH family.
Uniprot
H9J1E3
A0A212EM36
S4PTH5
A0A437BNR6
A0A212F363
A0A2H1V7M9
+ More
A0A2H1VEH3 A0A2A4JUL8 A0A2H1V6D2 A0A2A4JSV1 A0A2H1V6B1 A0A3S2PI34 A0A437BNT0 I4DKU7 A0A194PKP8 A0A194RJR2 H9J1E5 A0A0L7KWL6 A0A0L7RA08 H9J1E4 A0A154PJN3 A0A2S2NI66 A0A310SS75 J9K1E6 J9K8F8 A0A0J7KX96 A0A2H8TE51 A0A2J7PGB4 A0A087ZSJ3 A0A2J7PGB2 A0A0M8ZQI7 A0A232EVA2 A0A2A3E2X8 A0A0A9YSD7 A0A232EJH0 K7J9V9 A0A2J7QJF6 R4G3N5 A0A232EHL1 C3XV52 A0A067R7I0 A0A0C9RAJ3 A0A2K8JLG6 K7J4C8 K7J4B7 A0A1D2M8Y6 A0A3B4VQ68 A0A3P8WF49 A0A3B4WIG6 A0A2J7PGB5 A0A1B6IVS6 W5JQK0 A0A232FNC3 A0A3Q0GT80 A0A2U9AVS5 G3MT96 A0A023GLH1 A0A026W765 A0A182N906 A0A1H1C1Y4 A0A182J4J6 W5J577 A0A232F8K5 A0A158NM46 K7JBQ6 A0A151JU46 K1R2P0 A0A232EPX4 R4WU43 A0A023F9W2 C3XR56 K7FNP7 A0A2M9EKL2 A0A182FS73 A0A151J0Y7 A0A3L8D8X9 A0A023ERL2 A0A232FJG7 A0A182VW49 A0A084W0F9 K7IWB6 A0A3M1QFP4 A0A3Q2VR14 A0A151IBQ5 A0A3P9CLW3 A0A3Q2YSP6 A0A182QPL3 K7J9W0 A0A3P8ND07 A0A0W8DDY9 W2X762 W2QCP2 W2ZG68 A0A081AEM4 W2NIE3 V9FCM4 A0A195B527 A0A226E032
A0A2H1VEH3 A0A2A4JUL8 A0A2H1V6D2 A0A2A4JSV1 A0A2H1V6B1 A0A3S2PI34 A0A437BNT0 I4DKU7 A0A194PKP8 A0A194RJR2 H9J1E5 A0A0L7KWL6 A0A0L7RA08 H9J1E4 A0A154PJN3 A0A2S2NI66 A0A310SS75 J9K1E6 J9K8F8 A0A0J7KX96 A0A2H8TE51 A0A2J7PGB4 A0A087ZSJ3 A0A2J7PGB2 A0A0M8ZQI7 A0A232EVA2 A0A2A3E2X8 A0A0A9YSD7 A0A232EJH0 K7J9V9 A0A2J7QJF6 R4G3N5 A0A232EHL1 C3XV52 A0A067R7I0 A0A0C9RAJ3 A0A2K8JLG6 K7J4C8 K7J4B7 A0A1D2M8Y6 A0A3B4VQ68 A0A3P8WF49 A0A3B4WIG6 A0A2J7PGB5 A0A1B6IVS6 W5JQK0 A0A232FNC3 A0A3Q0GT80 A0A2U9AVS5 G3MT96 A0A023GLH1 A0A026W765 A0A182N906 A0A1H1C1Y4 A0A182J4J6 W5J577 A0A232F8K5 A0A158NM46 K7JBQ6 A0A151JU46 K1R2P0 A0A232EPX4 R4WU43 A0A023F9W2 C3XR56 K7FNP7 A0A2M9EKL2 A0A182FS73 A0A151J0Y7 A0A3L8D8X9 A0A023ERL2 A0A232FJG7 A0A182VW49 A0A084W0F9 K7IWB6 A0A3M1QFP4 A0A3Q2VR14 A0A151IBQ5 A0A3P9CLW3 A0A3Q2YSP6 A0A182QPL3 K7J9W0 A0A3P8ND07 A0A0W8DDY9 W2X762 W2QCP2 W2ZG68 A0A081AEM4 W2NIE3 V9FCM4 A0A195B527 A0A226E032
Pubmed
EMBL
BABH01023869
AGBW02013924
OWR42554.1
GAIX01012028
JAA80532.1
RSAL01000026
+ More
RVE52041.1 AGBW02010611 OWR48179.1 ODYU01000913 SOQ36392.1 ODYU01002133 SOQ39238.1 NWSH01000641 PCG75092.1 SOQ36391.1 PCG75091.1 ODYU01000912 SOQ36390.1 RVE52040.1 RVE52039.1 AK401915 BAM18537.1 KQ459602 KPI93578.1 KQ460106 KPJ17792.1 BABH01023867 JTDY01004982 KOB67515.1 KQ414619 KOC67707.1 BABH01023868 KQ434912 KZC11420.1 GGMR01004199 MBY16818.1 KQ760974 OAD58555.1 ABLF02039477 ABLF02034876 LBMM01002383 KMQ94899.1 GFXV01000566 MBW12371.1 NEVH01025635 PNF15371.1 PNF15370.1 KQ435903 KOX69065.1 NNAY01002030 OXU22265.1 KZ288446 PBC25616.1 GBHO01011144 GBHO01011140 GBHO01011137 GBRD01012537 GBRD01012536 GBRD01000907 GBRD01000906 GBRD01000905 GDHC01017642 JAG32460.1 JAG32464.1 JAG32467.1 JAG53287.1 JAQ00987.1 NNAY01004007 OXU18506.1 AAZX01002793 AAZX01017422 NEVH01013556 PNF28723.1 GAHY01001506 JAA76004.1 NNAY01004499 OXU17811.1 GG666468 EEN67978.1 KK852824 KDR15455.1 GBYB01000894 GBYB01005290 GBYB01005291 JAG70661.1 JAG75057.1 JAG75058.1 MF683286 ATU82427.1 AAZX01007303 AAZX01006389 LJIJ01002632 ODM89447.1 PNF15373.1 GECU01016659 JAS91047.1 ADMH02000745 ETN65185.1 NNAY01000004 OXU32214.1 CP026243 AWO95711.1 JO845097 AEO36714.1 GBBM01001753 JAC33665.1 KK107390 EZA51461.1 FNLD01000001 SDQ58174.1 ADMH02002189 ETN57899.1 NNAY01000718 OXU26820.1 ADTU01020239 AAZX01004663 AAZX01005711 AAZX01011636 KQ981842 KYN35001.1 JH815821 EKC40008.1 NNAY01002872 OXU20403.1 AK418237 BAN21452.1 GBBI01000431 JAC18281.1 GG666456 EEN69174.1 AGCU01191735 AGCU01191736 AGCU01191737 NWQS01000021 PJK11756.1 KQ980586 KYN15233.1 QOIP01000011 RLU16990.1 GAPW01002282 JAC11316.1 NNAY01000110 OXU30884.1 ATLV01019121 KE525262 KFB43703.1 RFIA01000250 RMF78724.1 KQ978079 KYM97157.1 AXCN02001403 LNFP01000287 KUF94579.1 ANIX01001491 ETP18362.1 KI669581 ETN10927.1 ANIY01001549 ETP46273.1 ANJA01001394 ETO77335.1 KI685887 KI672486 KI679208 KI692433 ETK88505.1 ETL41903.1 ETL95057.1 ETM48290.1 ANIZ01001242 ANIZ01001241 ETI48533.1 ETI48561.1 KQ976604 KYM79372.1 LNIX01000009 OXA50404.1
RVE52041.1 AGBW02010611 OWR48179.1 ODYU01000913 SOQ36392.1 ODYU01002133 SOQ39238.1 NWSH01000641 PCG75092.1 SOQ36391.1 PCG75091.1 ODYU01000912 SOQ36390.1 RVE52040.1 RVE52039.1 AK401915 BAM18537.1 KQ459602 KPI93578.1 KQ460106 KPJ17792.1 BABH01023867 JTDY01004982 KOB67515.1 KQ414619 KOC67707.1 BABH01023868 KQ434912 KZC11420.1 GGMR01004199 MBY16818.1 KQ760974 OAD58555.1 ABLF02039477 ABLF02034876 LBMM01002383 KMQ94899.1 GFXV01000566 MBW12371.1 NEVH01025635 PNF15371.1 PNF15370.1 KQ435903 KOX69065.1 NNAY01002030 OXU22265.1 KZ288446 PBC25616.1 GBHO01011144 GBHO01011140 GBHO01011137 GBRD01012537 GBRD01012536 GBRD01000907 GBRD01000906 GBRD01000905 GDHC01017642 JAG32460.1 JAG32464.1 JAG32467.1 JAG53287.1 JAQ00987.1 NNAY01004007 OXU18506.1 AAZX01002793 AAZX01017422 NEVH01013556 PNF28723.1 GAHY01001506 JAA76004.1 NNAY01004499 OXU17811.1 GG666468 EEN67978.1 KK852824 KDR15455.1 GBYB01000894 GBYB01005290 GBYB01005291 JAG70661.1 JAG75057.1 JAG75058.1 MF683286 ATU82427.1 AAZX01007303 AAZX01006389 LJIJ01002632 ODM89447.1 PNF15373.1 GECU01016659 JAS91047.1 ADMH02000745 ETN65185.1 NNAY01000004 OXU32214.1 CP026243 AWO95711.1 JO845097 AEO36714.1 GBBM01001753 JAC33665.1 KK107390 EZA51461.1 FNLD01000001 SDQ58174.1 ADMH02002189 ETN57899.1 NNAY01000718 OXU26820.1 ADTU01020239 AAZX01004663 AAZX01005711 AAZX01011636 KQ981842 KYN35001.1 JH815821 EKC40008.1 NNAY01002872 OXU20403.1 AK418237 BAN21452.1 GBBI01000431 JAC18281.1 GG666456 EEN69174.1 AGCU01191735 AGCU01191736 AGCU01191737 NWQS01000021 PJK11756.1 KQ980586 KYN15233.1 QOIP01000011 RLU16990.1 GAPW01002282 JAC11316.1 NNAY01000110 OXU30884.1 ATLV01019121 KE525262 KFB43703.1 RFIA01000250 RMF78724.1 KQ978079 KYM97157.1 AXCN02001403 LNFP01000287 KUF94579.1 ANIX01001491 ETP18362.1 KI669581 ETN10927.1 ANIY01001549 ETP46273.1 ANJA01001394 ETO77335.1 KI685887 KI672486 KI679208 KI692433 ETK88505.1 ETL41903.1 ETL95057.1 ETM48290.1 ANIZ01001242 ANIZ01001241 ETI48533.1 ETI48561.1 KQ976604 KYM79372.1 LNIX01000009 OXA50404.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000037510 UP000053825 UP000076502 UP000007819 UP000036403 UP000235965 UP000005203 UP000053105 UP000215335 UP000242457 UP000002358 UP000001554 UP000027135 UP000094527 UP000261420 UP000265120 UP000261360 UP000000673 UP000189705 UP000246464 UP000053097 UP000075884 UP000199049 UP000075880 UP000005205 UP000078541 UP000005408 UP000007267 UP000231561 UP000069272 UP000078492 UP000279307 UP000075920 UP000030765 UP000269435 UP000264840 UP000078542 UP000265160 UP000264820 UP000075886 UP000265100 UP000018958 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054423 UP000054532 UP000018721 UP000078540 UP000198287
UP000037510 UP000053825 UP000076502 UP000007819 UP000036403 UP000235965 UP000005203 UP000053105 UP000215335 UP000242457 UP000002358 UP000001554 UP000027135 UP000094527 UP000261420 UP000265120 UP000261360 UP000000673 UP000189705 UP000246464 UP000053097 UP000075884 UP000199049 UP000075880 UP000005205 UP000078541 UP000005408 UP000007267 UP000231561 UP000069272 UP000078492 UP000279307 UP000075920 UP000030765 UP000269435 UP000264840 UP000078542 UP000265160 UP000264820 UP000075886 UP000265100 UP000018958 UP000018817 UP000018948 UP000028582 UP000053236 UP000053864 UP000054423 UP000054532 UP000018721 UP000078540 UP000198287
Pfam
Interpro
IPR001910
Inosine/uridine_hydrolase_dom
+ More
IPR036452 Ribo_hydro-like
IPR034113 SCP_GAPR1-like
IPR001283 CRISP-related
IPR035940 CAP_sf
IPR018244 Allrgn_V5/Tpx1_CS
IPR014044 CAP_domain
IPR007379 Tim44-like_dom
IPR032710 NTF2-like_dom_sf
IPR039544 Tim44-like
IPR006554 Helicase-like_DEXD_c2
IPR027417 P-loop_NTPase
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR028331 CHL1/DDX11
IPR010614 DEAD_2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR015910 I/U_nuclsd_hydro_CS
IPR000719 Prot_kinase_dom
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR036137 Focal_adhe_kin_target_dom_sf
IPR019749 Band_41_domain
IPR020635 Tyr_kinase_cat_dom
IPR005189 Focal_adhesion_kin_target_dom
IPR035963 FERM_2
IPR041784 FAK1/PYK2_FERM_C
IPR008266 Tyr_kinase_AS
IPR041390 FADK_N
IPR019748 FERM_central
IPR017441 Protein_kinase_ATP_BS
IPR011993 PH-like_dom_sf
IPR011009 Kinase-like_dom_sf
IPR030610 PTK2B
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR023186 IUNH
IPR036452 Ribo_hydro-like
IPR034113 SCP_GAPR1-like
IPR001283 CRISP-related
IPR035940 CAP_sf
IPR018244 Allrgn_V5/Tpx1_CS
IPR014044 CAP_domain
IPR007379 Tim44-like_dom
IPR032710 NTF2-like_dom_sf
IPR039544 Tim44-like
IPR006554 Helicase-like_DEXD_c2
IPR027417 P-loop_NTPase
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR028331 CHL1/DDX11
IPR010614 DEAD_2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR015910 I/U_nuclsd_hydro_CS
IPR000719 Prot_kinase_dom
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR036137 Focal_adhe_kin_target_dom_sf
IPR019749 Band_41_domain
IPR020635 Tyr_kinase_cat_dom
IPR005189 Focal_adhesion_kin_target_dom
IPR035963 FERM_2
IPR041784 FAK1/PYK2_FERM_C
IPR008266 Tyr_kinase_AS
IPR041390 FADK_N
IPR019748 FERM_central
IPR017441 Protein_kinase_ATP_BS
IPR011993 PH-like_dom_sf
IPR011009 Kinase-like_dom_sf
IPR030610 PTK2B
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000299 FERM_domain
IPR029071 Ubiquitin-like_domsf
IPR023186 IUNH
SUPFAM
Gene 3D
ProteinModelPortal
H9J1E3
A0A212EM36
S4PTH5
A0A437BNR6
A0A212F363
A0A2H1V7M9
+ More
A0A2H1VEH3 A0A2A4JUL8 A0A2H1V6D2 A0A2A4JSV1 A0A2H1V6B1 A0A3S2PI34 A0A437BNT0 I4DKU7 A0A194PKP8 A0A194RJR2 H9J1E5 A0A0L7KWL6 A0A0L7RA08 H9J1E4 A0A154PJN3 A0A2S2NI66 A0A310SS75 J9K1E6 J9K8F8 A0A0J7KX96 A0A2H8TE51 A0A2J7PGB4 A0A087ZSJ3 A0A2J7PGB2 A0A0M8ZQI7 A0A232EVA2 A0A2A3E2X8 A0A0A9YSD7 A0A232EJH0 K7J9V9 A0A2J7QJF6 R4G3N5 A0A232EHL1 C3XV52 A0A067R7I0 A0A0C9RAJ3 A0A2K8JLG6 K7J4C8 K7J4B7 A0A1D2M8Y6 A0A3B4VQ68 A0A3P8WF49 A0A3B4WIG6 A0A2J7PGB5 A0A1B6IVS6 W5JQK0 A0A232FNC3 A0A3Q0GT80 A0A2U9AVS5 G3MT96 A0A023GLH1 A0A026W765 A0A182N906 A0A1H1C1Y4 A0A182J4J6 W5J577 A0A232F8K5 A0A158NM46 K7JBQ6 A0A151JU46 K1R2P0 A0A232EPX4 R4WU43 A0A023F9W2 C3XR56 K7FNP7 A0A2M9EKL2 A0A182FS73 A0A151J0Y7 A0A3L8D8X9 A0A023ERL2 A0A232FJG7 A0A182VW49 A0A084W0F9 K7IWB6 A0A3M1QFP4 A0A3Q2VR14 A0A151IBQ5 A0A3P9CLW3 A0A3Q2YSP6 A0A182QPL3 K7J9W0 A0A3P8ND07 A0A0W8DDY9 W2X762 W2QCP2 W2ZG68 A0A081AEM4 W2NIE3 V9FCM4 A0A195B527 A0A226E032
A0A2H1VEH3 A0A2A4JUL8 A0A2H1V6D2 A0A2A4JSV1 A0A2H1V6B1 A0A3S2PI34 A0A437BNT0 I4DKU7 A0A194PKP8 A0A194RJR2 H9J1E5 A0A0L7KWL6 A0A0L7RA08 H9J1E4 A0A154PJN3 A0A2S2NI66 A0A310SS75 J9K1E6 J9K8F8 A0A0J7KX96 A0A2H8TE51 A0A2J7PGB4 A0A087ZSJ3 A0A2J7PGB2 A0A0M8ZQI7 A0A232EVA2 A0A2A3E2X8 A0A0A9YSD7 A0A232EJH0 K7J9V9 A0A2J7QJF6 R4G3N5 A0A232EHL1 C3XV52 A0A067R7I0 A0A0C9RAJ3 A0A2K8JLG6 K7J4C8 K7J4B7 A0A1D2M8Y6 A0A3B4VQ68 A0A3P8WF49 A0A3B4WIG6 A0A2J7PGB5 A0A1B6IVS6 W5JQK0 A0A232FNC3 A0A3Q0GT80 A0A2U9AVS5 G3MT96 A0A023GLH1 A0A026W765 A0A182N906 A0A1H1C1Y4 A0A182J4J6 W5J577 A0A232F8K5 A0A158NM46 K7JBQ6 A0A151JU46 K1R2P0 A0A232EPX4 R4WU43 A0A023F9W2 C3XR56 K7FNP7 A0A2M9EKL2 A0A182FS73 A0A151J0Y7 A0A3L8D8X9 A0A023ERL2 A0A232FJG7 A0A182VW49 A0A084W0F9 K7IWB6 A0A3M1QFP4 A0A3Q2VR14 A0A151IBQ5 A0A3P9CLW3 A0A3Q2YSP6 A0A182QPL3 K7J9W0 A0A3P8ND07 A0A0W8DDY9 W2X762 W2QCP2 W2ZG68 A0A081AEM4 W2NIE3 V9FCM4 A0A195B527 A0A226E032
PDB
3T8I
E-value=1.58711e-24,
Score=279
Ontologies
GO
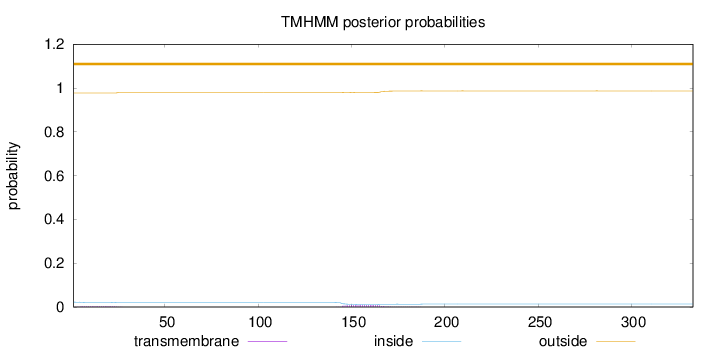
Topology
Subcellular location
Nucleus
SignalP
Position: 1 - 22,
Likelihood: 0.977017
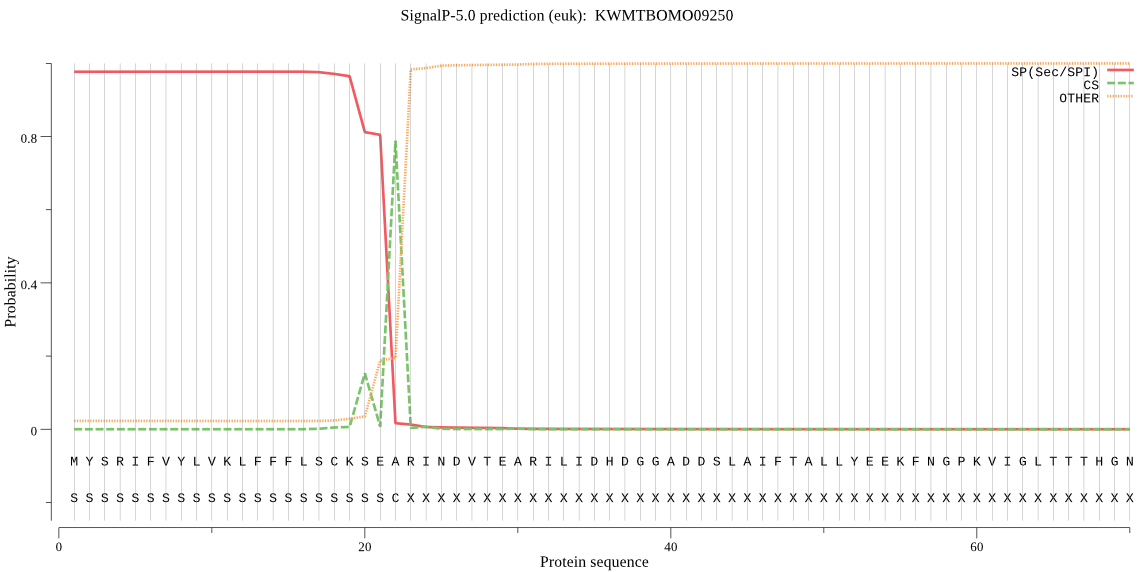
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.260450000000001
Exp number, first 60 AAs:
0.05948
Total prob of N-in:
0.02242
outside
1 - 333
Population Genetic Test Statistics
Pi
252.90522
Theta
172.17183
Tajima's D
2.164527
CLR
0.159016
CSRT
0.902954852257387
Interpretation
Uncertain
