Gene
KWMTBOMO09249
Annotation
PREDICTED:_uncharacterized_protein_C1683.06c-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.725 Extracellular Reliability : 1.311
Sequence
CDS
ATGACAGGAATTAAGAGGAAATTCGTGATCGACAATGACGCTGGCGGTGATGATGCTATGGCCATATTCTTGGCCGCTCTGTTCGAGAAGTATCATGCCGGTCCTCAATTAATTGGAGTTACGACAAGCAACGGTAACACCAACGAAGACAACGTTTCTTACAATAACCAGAGAATACTGAAGGTCGCCAAAAGACAGGACGTGCCAATTTATAGAGGTTCGAAATCCTCTTTGGTGAAAACCCCTGAAATCACGGACTACTTCGGTAAAGACGGCCTCGGAGACAGTGGGGATGTCTACCCAGATCTGGTACCGCCGCATACAGAAAATGCCGTCAACGCTCTTATACAATTGTCTAAGACACATGAAGGCAACCTTACCATCATCACTATAGGCGCGCTAACCAACTTAGCTTTGGCCATTAAAACGGACCCCGCCTTTCTCGGCAGACTCGCACACGTCTACATCGGAGCCGGACACATACATACCGAAGAATACCCGACAGCAGAGTTCAACGCTCACATGGATGTTGAAGCGTACCACGTCGTGACCGAAAACGCGAATCCAGAGAAAGTTACAATATTCCCCTTCTCACAGGTGCAGAAATATTGTAACTTTAGCCGGGAGTGGAGGATAAACGTTCTCGGCCGGATTTAG
Protein
MTGIKRKFVIDNDAGGDDAMAIFLAALFEKYHAGPQLIGVTTSNGNTNEDNVSYNNQRILKVAKRQDVPIYRGSKSSLVKTPEITDYFGKDGLGDSGDVYPDLVPPHTENAVNALIQLSKTHEGNLTIITIGALTNLALAIKTDPAFLGRLAHVYIGAGHIHTEEYPTAEFNAHMDVEAYHVVTENANPEKVTIFPFSQVQKYCNFSREWRINVLGRI
Summary
Similarity
Belongs to the CRISP family.
Uniprot
A0A2A4JSV1
A0A2H1V6B1
A0A212F363
S4PTH5
I4DKU7
A0A437BNT0
+ More
A0A194PKP8 A0A3S2PI34 A0A194RJR2 H9J1E4 H9J1E5 A0A212EM36 A0A437BNR6 H9J1E3 A0A2H1V6D2 A0A2H1V7M9 A0A2H1VEH3 A0A2A4JUL8 A0A0L7KWL6 A0A2J7QJF6 A0A2J7PGB2 A0A232EVA2 A0A2J7PGB4 A0A067R7I0 A0A182M9Y1 A0A1W4ZYG1 A0A182R4F1 A0A182QPL3 K7J4C8 A0A182YL46 A0A3B1JLF6 A0A0P4Z7T0 A0A182N906 A0A0P4YRB0 A0A0N8C0V8 A0A162R495 A0A182WED0 A0A0P5H8B6 E9HSD0 A0A2D0RVU6 K7J9V9 A0A182V527 A0A0N8A1D7 A0A0N7ZIB0 V9KSU3 A0A232EPX4 A0A2B4RPW5 A0A182TTK6 A0A3B4EK92 K7J9W1 A0A3B3HYZ7 A0A3P9KZ16 K7J4B7 A0A3P9JCS3 A0A1S4H8C1 A0A182L4K9 A0A182HH40 A0A3B3RFI1 Q7PZ48 C3XV52 A0A232EJH0 A0A1I8JA73 A0A1I8J8V7 V4A517 A0A0P6HC41 A0A3P9KYP5 A0A182X6N5 E1ZUY1 A0A3P9MBH2 A0A3M6TBA3 W4Z6M6 A0A182P6Y1 A0A0M8ZQI7 A0A3Q0S0V8 A0A0A9W0Q8 A0A3P9CLW3 A0A2U9AVS9 A0A232F8K5 A0A151JU46 A0A0P7U892 A0A3Q1H5H0 A0A3L7ACX1 A0A3Q2YSP6 A0A3B4HBK2 A0A3P9CLT6 A0A232EHL1 A0A3B4XIJ0 A0A2J7PGB5 A0A0L7RA08 A0A401P6R6 A0A401SBJ2 A0A3P8ND07 A0A2U9C6R0 A0A0P6CRB8 W5MPL0 F4WHN9 A0A3Q2VR14 A0A3B4HDL0 A7S2K9 A0A1D1V4P4
A0A194PKP8 A0A3S2PI34 A0A194RJR2 H9J1E4 H9J1E5 A0A212EM36 A0A437BNR6 H9J1E3 A0A2H1V6D2 A0A2H1V7M9 A0A2H1VEH3 A0A2A4JUL8 A0A0L7KWL6 A0A2J7QJF6 A0A2J7PGB2 A0A232EVA2 A0A2J7PGB4 A0A067R7I0 A0A182M9Y1 A0A1W4ZYG1 A0A182R4F1 A0A182QPL3 K7J4C8 A0A182YL46 A0A3B1JLF6 A0A0P4Z7T0 A0A182N906 A0A0P4YRB0 A0A0N8C0V8 A0A162R495 A0A182WED0 A0A0P5H8B6 E9HSD0 A0A2D0RVU6 K7J9V9 A0A182V527 A0A0N8A1D7 A0A0N7ZIB0 V9KSU3 A0A232EPX4 A0A2B4RPW5 A0A182TTK6 A0A3B4EK92 K7J9W1 A0A3B3HYZ7 A0A3P9KZ16 K7J4B7 A0A3P9JCS3 A0A1S4H8C1 A0A182L4K9 A0A182HH40 A0A3B3RFI1 Q7PZ48 C3XV52 A0A232EJH0 A0A1I8JA73 A0A1I8J8V7 V4A517 A0A0P6HC41 A0A3P9KYP5 A0A182X6N5 E1ZUY1 A0A3P9MBH2 A0A3M6TBA3 W4Z6M6 A0A182P6Y1 A0A0M8ZQI7 A0A3Q0S0V8 A0A0A9W0Q8 A0A3P9CLW3 A0A2U9AVS9 A0A232F8K5 A0A151JU46 A0A0P7U892 A0A3Q1H5H0 A0A3L7ACX1 A0A3Q2YSP6 A0A3B4HBK2 A0A3P9CLT6 A0A232EHL1 A0A3B4XIJ0 A0A2J7PGB5 A0A0L7RA08 A0A401P6R6 A0A401SBJ2 A0A3P8ND07 A0A2U9C6R0 A0A0P6CRB8 W5MPL0 F4WHN9 A0A3Q2VR14 A0A3B4HDL0 A7S2K9 A0A1D1V4P4
Pubmed
EMBL
NWSH01000641
PCG75091.1
ODYU01000912
SOQ36390.1
AGBW02010611
OWR48179.1
+ More
GAIX01012028 JAA80532.1 AK401915 BAM18537.1 RSAL01000026 RVE52039.1 KQ459602 KPI93578.1 RVE52040.1 KQ460106 KPJ17792.1 BABH01023867 BABH01023868 BABH01023869 AGBW02013924 OWR42554.1 RVE52041.1 ODYU01000913 SOQ36391.1 SOQ36392.1 ODYU01002133 SOQ39238.1 PCG75092.1 JTDY01004982 KOB67515.1 NEVH01013556 PNF28723.1 NEVH01025635 PNF15370.1 NNAY01002030 OXU22265.1 PNF15371.1 KK852824 KDR15455.1 AXCM01000780 AXCN02001403 AAZX01007303 GDIP01216639 JAJ06763.1 GDIP01224935 JAI98466.1 GDIQ01125795 JAL25931.1 LRGB01000243 KZS20217.1 GDIQ01229952 JAK21773.1 GL732749 EFX65347.1 AAZX01002793 AAZX01017422 GDIP01192041 JAJ31361.1 GDIP01242657 JAI80744.1 JW868927 AFP01445.1 NNAY01002872 OXU20403.1 LSMT01000316 PFX20464.1 AAZX01013758 AAZX01006389 AAAB01008986 APCN01002384 EAA00053.3 GG666468 EEN67978.1 NNAY01004007 OXU18506.1 NIVC01000958 PAA74291.1 NIVC01000712 PAA78034.1 KB201205 ESO98993.1 GDIQ01021770 JAN72967.1 GL434335 EFN74979.1 RCHS01003977 RMX38619.1 AAGJ04152716 KQ435903 KOX69065.1 GBHO01043586 GBHO01043585 GBRD01016007 GBRD01008012 GBRD01008011 GBRD01008009 GDHC01015347 JAG00018.1 JAG00019.1 JAG49819.1 JAQ03282.1 CP026243 AWO95710.1 NNAY01000718 OXU26820.1 KQ981842 KYN35001.1 JARO02007536 KPP63836.1 RCUX01000002 RLP77578.1 NNAY01004499 OXU17811.1 PNF15373.1 KQ414619 KOC67707.1 BFAA01003142 GCB68831.1 BEZZ01000176 GCC27766.1 CP026254 AWP11416.1 GDIQ01088659 JAN06078.1 AHAT01003407 GL888167 EGI66246.1 DS469569 EDO42061.1 BDGG01000003 GAU95890.1
GAIX01012028 JAA80532.1 AK401915 BAM18537.1 RSAL01000026 RVE52039.1 KQ459602 KPI93578.1 RVE52040.1 KQ460106 KPJ17792.1 BABH01023867 BABH01023868 BABH01023869 AGBW02013924 OWR42554.1 RVE52041.1 ODYU01000913 SOQ36391.1 SOQ36392.1 ODYU01002133 SOQ39238.1 PCG75092.1 JTDY01004982 KOB67515.1 NEVH01013556 PNF28723.1 NEVH01025635 PNF15370.1 NNAY01002030 OXU22265.1 PNF15371.1 KK852824 KDR15455.1 AXCM01000780 AXCN02001403 AAZX01007303 GDIP01216639 JAJ06763.1 GDIP01224935 JAI98466.1 GDIQ01125795 JAL25931.1 LRGB01000243 KZS20217.1 GDIQ01229952 JAK21773.1 GL732749 EFX65347.1 AAZX01002793 AAZX01017422 GDIP01192041 JAJ31361.1 GDIP01242657 JAI80744.1 JW868927 AFP01445.1 NNAY01002872 OXU20403.1 LSMT01000316 PFX20464.1 AAZX01013758 AAZX01006389 AAAB01008986 APCN01002384 EAA00053.3 GG666468 EEN67978.1 NNAY01004007 OXU18506.1 NIVC01000958 PAA74291.1 NIVC01000712 PAA78034.1 KB201205 ESO98993.1 GDIQ01021770 JAN72967.1 GL434335 EFN74979.1 RCHS01003977 RMX38619.1 AAGJ04152716 KQ435903 KOX69065.1 GBHO01043586 GBHO01043585 GBRD01016007 GBRD01008012 GBRD01008011 GBRD01008009 GDHC01015347 JAG00018.1 JAG00019.1 JAG49819.1 JAQ03282.1 CP026243 AWO95710.1 NNAY01000718 OXU26820.1 KQ981842 KYN35001.1 JARO02007536 KPP63836.1 RCUX01000002 RLP77578.1 NNAY01004499 OXU17811.1 PNF15373.1 KQ414619 KOC67707.1 BFAA01003142 GCB68831.1 BEZZ01000176 GCC27766.1 CP026254 AWP11416.1 GDIQ01088659 JAN06078.1 AHAT01003407 GL888167 EGI66246.1 DS469569 EDO42061.1 BDGG01000003 GAU95890.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000005204
+ More
UP000037510 UP000235965 UP000215335 UP000027135 UP000075883 UP000192224 UP000075900 UP000075886 UP000002358 UP000076408 UP000018467 UP000075884 UP000076858 UP000075920 UP000000305 UP000221080 UP000075903 UP000225706 UP000075902 UP000261440 UP000001038 UP000265180 UP000265200 UP000075882 UP000075840 UP000261540 UP000007062 UP000001554 UP000095280 UP000215902 UP000030746 UP000076407 UP000000311 UP000275408 UP000007110 UP000075885 UP000053105 UP000261340 UP000265160 UP000246464 UP000078541 UP000034805 UP000265040 UP000272503 UP000264820 UP000261460 UP000261360 UP000053825 UP000288216 UP000287033 UP000265100 UP000018468 UP000007755 UP000264840 UP000001593 UP000186922
UP000037510 UP000235965 UP000215335 UP000027135 UP000075883 UP000192224 UP000075900 UP000075886 UP000002358 UP000076408 UP000018467 UP000075884 UP000076858 UP000075920 UP000000305 UP000221080 UP000075903 UP000225706 UP000075902 UP000261440 UP000001038 UP000265180 UP000265200 UP000075882 UP000075840 UP000261540 UP000007062 UP000001554 UP000095280 UP000215902 UP000030746 UP000076407 UP000000311 UP000275408 UP000007110 UP000075885 UP000053105 UP000261340 UP000265160 UP000246464 UP000078541 UP000034805 UP000265040 UP000272503 UP000264820 UP000261460 UP000261360 UP000053825 UP000288216 UP000287033 UP000265100 UP000018468 UP000007755 UP000264840 UP000001593 UP000186922
Interpro
IPR036452
Ribo_hydro-like
+ More
IPR001910 Inosine/uridine_hydrolase_dom
IPR034113 SCP_GAPR1-like
IPR001283 CRISP-related
IPR035940 CAP_sf
IPR018244 Allrgn_V5/Tpx1_CS
IPR014044 CAP_domain
IPR006554 Helicase-like_DEXD_c2
IPR027417 P-loop_NTPase
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR028331 CHL1/DDX11
IPR010614 DEAD_2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR007379 Tim44-like_dom
IPR032710 NTF2-like_dom_sf
IPR039544 Tim44-like
IPR015910 I/U_nuclsd_hydro_CS
IPR001910 Inosine/uridine_hydrolase_dom
IPR034113 SCP_GAPR1-like
IPR001283 CRISP-related
IPR035940 CAP_sf
IPR018244 Allrgn_V5/Tpx1_CS
IPR014044 CAP_domain
IPR006554 Helicase-like_DEXD_c2
IPR027417 P-loop_NTPase
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR028331 CHL1/DDX11
IPR010614 DEAD_2
IPR013020 Rad3/Chl1-like
IPR006555 ATP-dep_Helicase_C
IPR007379 Tim44-like_dom
IPR032710 NTF2-like_dom_sf
IPR039544 Tim44-like
IPR015910 I/U_nuclsd_hydro_CS
Gene 3D
ProteinModelPortal
A0A2A4JSV1
A0A2H1V6B1
A0A212F363
S4PTH5
I4DKU7
A0A437BNT0
+ More
A0A194PKP8 A0A3S2PI34 A0A194RJR2 H9J1E4 H9J1E5 A0A212EM36 A0A437BNR6 H9J1E3 A0A2H1V6D2 A0A2H1V7M9 A0A2H1VEH3 A0A2A4JUL8 A0A0L7KWL6 A0A2J7QJF6 A0A2J7PGB2 A0A232EVA2 A0A2J7PGB4 A0A067R7I0 A0A182M9Y1 A0A1W4ZYG1 A0A182R4F1 A0A182QPL3 K7J4C8 A0A182YL46 A0A3B1JLF6 A0A0P4Z7T0 A0A182N906 A0A0P4YRB0 A0A0N8C0V8 A0A162R495 A0A182WED0 A0A0P5H8B6 E9HSD0 A0A2D0RVU6 K7J9V9 A0A182V527 A0A0N8A1D7 A0A0N7ZIB0 V9KSU3 A0A232EPX4 A0A2B4RPW5 A0A182TTK6 A0A3B4EK92 K7J9W1 A0A3B3HYZ7 A0A3P9KZ16 K7J4B7 A0A3P9JCS3 A0A1S4H8C1 A0A182L4K9 A0A182HH40 A0A3B3RFI1 Q7PZ48 C3XV52 A0A232EJH0 A0A1I8JA73 A0A1I8J8V7 V4A517 A0A0P6HC41 A0A3P9KYP5 A0A182X6N5 E1ZUY1 A0A3P9MBH2 A0A3M6TBA3 W4Z6M6 A0A182P6Y1 A0A0M8ZQI7 A0A3Q0S0V8 A0A0A9W0Q8 A0A3P9CLW3 A0A2U9AVS9 A0A232F8K5 A0A151JU46 A0A0P7U892 A0A3Q1H5H0 A0A3L7ACX1 A0A3Q2YSP6 A0A3B4HBK2 A0A3P9CLT6 A0A232EHL1 A0A3B4XIJ0 A0A2J7PGB5 A0A0L7RA08 A0A401P6R6 A0A401SBJ2 A0A3P8ND07 A0A2U9C6R0 A0A0P6CRB8 W5MPL0 F4WHN9 A0A3Q2VR14 A0A3B4HDL0 A7S2K9 A0A1D1V4P4
A0A194PKP8 A0A3S2PI34 A0A194RJR2 H9J1E4 H9J1E5 A0A212EM36 A0A437BNR6 H9J1E3 A0A2H1V6D2 A0A2H1V7M9 A0A2H1VEH3 A0A2A4JUL8 A0A0L7KWL6 A0A2J7QJF6 A0A2J7PGB2 A0A232EVA2 A0A2J7PGB4 A0A067R7I0 A0A182M9Y1 A0A1W4ZYG1 A0A182R4F1 A0A182QPL3 K7J4C8 A0A182YL46 A0A3B1JLF6 A0A0P4Z7T0 A0A182N906 A0A0P4YRB0 A0A0N8C0V8 A0A162R495 A0A182WED0 A0A0P5H8B6 E9HSD0 A0A2D0RVU6 K7J9V9 A0A182V527 A0A0N8A1D7 A0A0N7ZIB0 V9KSU3 A0A232EPX4 A0A2B4RPW5 A0A182TTK6 A0A3B4EK92 K7J9W1 A0A3B3HYZ7 A0A3P9KZ16 K7J4B7 A0A3P9JCS3 A0A1S4H8C1 A0A182L4K9 A0A182HH40 A0A3B3RFI1 Q7PZ48 C3XV52 A0A232EJH0 A0A1I8JA73 A0A1I8J8V7 V4A517 A0A0P6HC41 A0A3P9KYP5 A0A182X6N5 E1ZUY1 A0A3P9MBH2 A0A3M6TBA3 W4Z6M6 A0A182P6Y1 A0A0M8ZQI7 A0A3Q0S0V8 A0A0A9W0Q8 A0A3P9CLW3 A0A2U9AVS9 A0A232F8K5 A0A151JU46 A0A0P7U892 A0A3Q1H5H0 A0A3L7ACX1 A0A3Q2YSP6 A0A3B4HBK2 A0A3P9CLT6 A0A232EHL1 A0A3B4XIJ0 A0A2J7PGB5 A0A0L7RA08 A0A401P6R6 A0A401SBJ2 A0A3P8ND07 A0A2U9C6R0 A0A0P6CRB8 W5MPL0 F4WHN9 A0A3Q2VR14 A0A3B4HDL0 A7S2K9 A0A1D1V4P4
PDB
3T8I
E-value=7.2015e-22,
Score=253
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
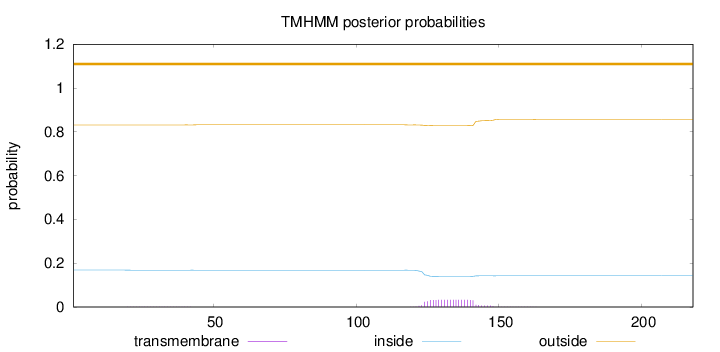
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.64292
Exp number, first 60 AAs:
0.01147
Total prob of N-in:
0.16848
outside
1 - 218
Population Genetic Test Statistics
Pi
303.281165
Theta
165.045749
Tajima's D
3.748218
CLR
0.256867
CSRT
0.99750012499375
Interpretation
Uncertain
