Gene
KWMTBOMO09236 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003335
Annotation
PREDICTED:_proteasome_subunit_beta_type-6_[Papilio_machaon]
Full name
Proteasome subunit beta
Location in the cell
Cytoplasmic Reliability : 1.681
Sequence
CDS
ATGGCAACATCAGTGATCAATACCTACTCTGATCCTTGCCTACCGGAGTGGATGAGCGCCCCTCATAGTACGGGAACTTCAATCATGGCATGCGAGTTTGATGGTGGAGTCGTCATTGGAGCTGATTCCCGTACCACAACAGGCGCTTACATCGCCAATCGAGTCACCGACAAGCTGACGAAAATCACTGACCAGATATACTGCTGCCGCTCAGGATCCGCTGCCGACACTCAAGCCATAGCTGATATTGTAACATATCATTTGAACTTCCACAAGATGGAATTAGGAGATCCACCACTAGTGGAAACTGCCGCAGCGATATTCCGTGAGCTCTGCTACAACTATCGTGACTCATTGATGGCTGGTATTTTAGTTGCTGGATGGGACAAAAAGAAAGGAGCTCAAATTTACTCTATACCTATTGGTGGTATGGTGCAGCGCCAAGCGGTGTCAATTGGAGGCTCCGGATCCTCTTACGTCTATGGCTACGTTGACGCCAATTTCAAGCCCAATATGACCAAGGAAGAAGCCAAAAAATTTGTTACGGACACACTGACATTGGCTATCCTTCGTGACGGCTCTTCTGGTGGTGTTGTCAGACTCGGAGTAATTACTGAAGCTGGTGTGGAACGCTCTGTTATTCTTGGAGACCAACTTCCCAAATTTTATGAAGGTTAA
Protein
MATSVINTYSDPCLPEWMSAPHSTGTSIMACEFDGGVVIGADSRTTTGAYIANRVTDKLTKITDQIYCCRSGSAADTQAIADIVTYHLNFHKMELGDPPLVETAAAIFRELCYNYRDSLMAGILVAGWDKKKGAQIYSIPIGGMVQRQAVSIGGSGSSYVYGYVDANFKPNMTKEEAKKFVTDTLTLAILRDGSSGGVVRLGVITEAGVERSVILGDQLPKFYEG
Summary
Description
The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH.
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
Similarity
Belongs to the peptidase T1B family.
Uniprot
H9J1E8
A0A2A4K9J9
A0A2H1VFD5
A0A194RJQ3
A0A194PKP4
A0A3S2LYP7
+ More
S4PHC9 I4DK39 A0A1E1WHA9 A0A212EXS6 A0A2P8YNA7 D6WT45 A0A0T6BB72 V5I9J7 K7J4Z1 A0A1Y1KTU2 J3JTW0 E1ZWG1 A0A2A3EMI3 V9II29 A0A0N0U515 A0A411G642 A0A232EYU2 A0A158NPP5 A0A0C9RFI6 A0A0L7R042 A0A195ESV4 A0A151X8H8 A0A195EM54 A0A1W4XAA1 A0A0J7L0J6 U4TYK1 N6TTR8 A0A026WTY8 A0A067R651 A0A226ERV9 U4U3C1 E9GZ78 A0A131XNR3 A0A224YG33 A0A131Z2F3 A0A023G7V6 A0A023FQ41 G3MRX7 A0A023FXX9 A0A0P5VFR9 A0A1E1X2R1 A0A0P6JVR0 A0A131XZ20 A0A154PEJ0 A0A0P5AUD2 A0A0P5WMP8 A0A0P4YC36 A0A0P5YL84 A0A0P5Y9B9 A0A151I8C8 E2BIK2 A0A2J7RR21 A0A023FNY4 A0A2R5LFX9 A0A1D2MXK3 A0A0P5BEP2 A0A0P4YZ09 K1QBT2 A0A3P9ADQ9 A9UWM7 A0A1Z5KYH6 F1RCM3 A0A1S3JWY0 B5X963 B5X8Y8 A0A060XEY9 A0A3B3RT78 C1BW52 A0A1W4Y7R5 A0A433UB52 A0A0P5RDI7 A0A3Q8QF21 B2GRB2 Q568V4 F4W7J5 B5XD66 O57332 R4UMP0 A0A444USC0 A0A060YSB5 A0A0P5ND39 A0A0K8RGM3 A0A1S3RIW7 A0A0B6ZXC6 A0A0P5N9Y1 W5USF9 A0A2D0QBT5 A0A151NTL8 A7RA95 C3ZIC3 A0A1P8DB30 A0A0L7KS96 A0A3P9DRT1 A0A3P8P4W9
S4PHC9 I4DK39 A0A1E1WHA9 A0A212EXS6 A0A2P8YNA7 D6WT45 A0A0T6BB72 V5I9J7 K7J4Z1 A0A1Y1KTU2 J3JTW0 E1ZWG1 A0A2A3EMI3 V9II29 A0A0N0U515 A0A411G642 A0A232EYU2 A0A158NPP5 A0A0C9RFI6 A0A0L7R042 A0A195ESV4 A0A151X8H8 A0A195EM54 A0A1W4XAA1 A0A0J7L0J6 U4TYK1 N6TTR8 A0A026WTY8 A0A067R651 A0A226ERV9 U4U3C1 E9GZ78 A0A131XNR3 A0A224YG33 A0A131Z2F3 A0A023G7V6 A0A023FQ41 G3MRX7 A0A023FXX9 A0A0P5VFR9 A0A1E1X2R1 A0A0P6JVR0 A0A131XZ20 A0A154PEJ0 A0A0P5AUD2 A0A0P5WMP8 A0A0P4YC36 A0A0P5YL84 A0A0P5Y9B9 A0A151I8C8 E2BIK2 A0A2J7RR21 A0A023FNY4 A0A2R5LFX9 A0A1D2MXK3 A0A0P5BEP2 A0A0P4YZ09 K1QBT2 A0A3P9ADQ9 A9UWM7 A0A1Z5KYH6 F1RCM3 A0A1S3JWY0 B5X963 B5X8Y8 A0A060XEY9 A0A3B3RT78 C1BW52 A0A1W4Y7R5 A0A433UB52 A0A0P5RDI7 A0A3Q8QF21 B2GRB2 Q568V4 F4W7J5 B5XD66 O57332 R4UMP0 A0A444USC0 A0A060YSB5 A0A0P5ND39 A0A0K8RGM3 A0A1S3RIW7 A0A0B6ZXC6 A0A0P5N9Y1 W5USF9 A0A2D0QBT5 A0A151NTL8 A7RA95 C3ZIC3 A0A1P8DB30 A0A0L7KS96 A0A3P9DRT1 A0A3P8P4W9
EC Number
3.4.25.1
Pubmed
19121390
26354079
23622113
22651552
22118469
29403074
+ More
18362917 19820115 20075255 28004739 22516182 20798317 28648823 21347285 23537049 24508170 30249741 24845553 21292972 28049606 28797301 26830274 22216098 28503490 27289101 22992520 25069045 18273011 28528879 23594743 20433749 24755649 29240929 21719571 9550404 23127152 22293439 18563158 26227816 25186727
18362917 19820115 20075255 28004739 22516182 20798317 28648823 21347285 23537049 24508170 30249741 24845553 21292972 28049606 28797301 26830274 22216098 28503490 27289101 22992520 25069045 18273011 28528879 23594743 20433749 24755649 29240929 21719571 9550404 23127152 22293439 18563158 26227816 25186727
EMBL
BABH01023829
NWSH01000012
PCG80887.1
ODYU01002066
SOQ39112.1
KQ460106
+ More
KPJ17782.1 KQ459602 KPI93573.1 RSAL01000112 RVE47034.1 GAIX01003322 JAA89238.1 AK401657 BAM18279.1 GDQN01004758 JAT86296.1 AGBW02011690 OWR46298.1 PYGN01000474 PSN45694.1 KQ971354 EFA05865.1 LJIG01002358 KRT84582.1 GALX01002815 JAB65651.1 GEZM01080221 JAV62287.1 BT126669 AEE61632.1 GL434830 EFN74473.1 KZ288217 PBC32369.1 JR048567 AEY60700.1 KQ435808 KOX72982.1 MH365582 QBB01391.1 NNAY01001584 OXU23525.1 ADTU01022697 GBYB01007155 JAG76922.1 KQ414672 KOC64220.1 KQ981986 KYN31236.1 KQ982409 KYQ56673.1 KQ978730 KYN28959.1 LBMM01001609 KMQ96019.1 KB631660 ERL85103.1 APGK01054753 KB741253 ENN71781.1 KK107105 QOIP01000011 EZA59530.1 RLU16727.1 KK852675 KDR18713.1 LNIX01000002 OXA59521.1 ERL85106.1 GL732577 EFX75100.1 GEFH01000783 JAP67798.1 GFPF01005501 MAA16647.1 GEDV01002948 JAP85609.1 GBBM01005504 JAC29914.1 GBBK01001058 JAC23424.1 JO844628 AEO36245.1 GBBL01001765 JAC25555.1 GDIP01166746 GDIP01100512 LRGB01001920 JAM03203.1 KZS10019.1 GFAC01005641 JAT93547.1 GDIQ01006896 JAN87841.1 GEFM01003313 JAP72483.1 KQ434889 KZC10251.1 GDIP01194005 JAJ29397.1 GDIP01085017 JAM18698.1 GDIP01229604 JAI93797.1 GDIP01057619 JAM46096.1 GDIP01061021 JAM42694.1 KQ978364 KYM94605.1 GL448516 EFN84449.1 NEVH01000611 PNF43275.1 GBBK01001021 JAC23461.1 GGLE01004209 MBY08335.1 LJIJ01000411 ODM97767.1 GDIP01189852 JAJ33550.1 GDIP01220802 JAJ02600.1 JH818665 EKC18961.1 CH991548 EDQ90240.1 GFJQ02007119 JAV99850.1 CU896563 CU929143 BT047582 ACI67383.1 BT047507 ACI67308.1 FR905301 CDQ78193.1 BT078831 ACO13255.1 RQTK01000019 RUS91039.1 GDIQ01108457 JAL43269.1 MH683009 AZI95695.1 BC165062 AAI65062.1 BC092699 AAH92699.1 GL887844 EGI69868.1 BT048985 ACI68786.1 AF032392 AAB87681.1 KC571886 AGM32385.1 SCEB01010584 RXM91063.1 FR913120 CDQ92344.1 GDIQ01144248 JAL07478.1 GADI01003790 JAA70018.1 HACG01026082 CEK72947.1 GDIQ01152140 JAK99585.1 JT417950 AHH42563.1 AKHW03002060 KYO40227.1 DQ279100 ABB86551.1 GG666627 EEN47648.1 KU351251 APU53803.1 JTDY01006283 KOB66142.1
KPJ17782.1 KQ459602 KPI93573.1 RSAL01000112 RVE47034.1 GAIX01003322 JAA89238.1 AK401657 BAM18279.1 GDQN01004758 JAT86296.1 AGBW02011690 OWR46298.1 PYGN01000474 PSN45694.1 KQ971354 EFA05865.1 LJIG01002358 KRT84582.1 GALX01002815 JAB65651.1 GEZM01080221 JAV62287.1 BT126669 AEE61632.1 GL434830 EFN74473.1 KZ288217 PBC32369.1 JR048567 AEY60700.1 KQ435808 KOX72982.1 MH365582 QBB01391.1 NNAY01001584 OXU23525.1 ADTU01022697 GBYB01007155 JAG76922.1 KQ414672 KOC64220.1 KQ981986 KYN31236.1 KQ982409 KYQ56673.1 KQ978730 KYN28959.1 LBMM01001609 KMQ96019.1 KB631660 ERL85103.1 APGK01054753 KB741253 ENN71781.1 KK107105 QOIP01000011 EZA59530.1 RLU16727.1 KK852675 KDR18713.1 LNIX01000002 OXA59521.1 ERL85106.1 GL732577 EFX75100.1 GEFH01000783 JAP67798.1 GFPF01005501 MAA16647.1 GEDV01002948 JAP85609.1 GBBM01005504 JAC29914.1 GBBK01001058 JAC23424.1 JO844628 AEO36245.1 GBBL01001765 JAC25555.1 GDIP01166746 GDIP01100512 LRGB01001920 JAM03203.1 KZS10019.1 GFAC01005641 JAT93547.1 GDIQ01006896 JAN87841.1 GEFM01003313 JAP72483.1 KQ434889 KZC10251.1 GDIP01194005 JAJ29397.1 GDIP01085017 JAM18698.1 GDIP01229604 JAI93797.1 GDIP01057619 JAM46096.1 GDIP01061021 JAM42694.1 KQ978364 KYM94605.1 GL448516 EFN84449.1 NEVH01000611 PNF43275.1 GBBK01001021 JAC23461.1 GGLE01004209 MBY08335.1 LJIJ01000411 ODM97767.1 GDIP01189852 JAJ33550.1 GDIP01220802 JAJ02600.1 JH818665 EKC18961.1 CH991548 EDQ90240.1 GFJQ02007119 JAV99850.1 CU896563 CU929143 BT047582 ACI67383.1 BT047507 ACI67308.1 FR905301 CDQ78193.1 BT078831 ACO13255.1 RQTK01000019 RUS91039.1 GDIQ01108457 JAL43269.1 MH683009 AZI95695.1 BC165062 AAI65062.1 BC092699 AAH92699.1 GL887844 EGI69868.1 BT048985 ACI68786.1 AF032392 AAB87681.1 KC571886 AGM32385.1 SCEB01010584 RXM91063.1 FR913120 CDQ92344.1 GDIQ01144248 JAL07478.1 GADI01003790 JAA70018.1 HACG01026082 CEK72947.1 GDIQ01152140 JAK99585.1 JT417950 AHH42563.1 AKHW03002060 KYO40227.1 DQ279100 ABB86551.1 GG666627 EEN47648.1 KU351251 APU53803.1 JTDY01006283 KOB66142.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000245037 UP000007266 UP000002358 UP000000311 UP000242457 UP000053105 UP000215335 UP000005205 UP000053825 UP000078541 UP000075809 UP000078492 UP000192223 UP000036403 UP000030742 UP000019118 UP000053097 UP000279307 UP000027135 UP000198287 UP000000305 UP000076858 UP000076502 UP000078542 UP000008237 UP000235965 UP000094527 UP000005408 UP000265140 UP000001357 UP000000437 UP000085678 UP000087266 UP000193380 UP000261540 UP000192224 UP000271974 UP000007755 UP000221080 UP000050525 UP000001554 UP000037510 UP000265160 UP000265100
UP000245037 UP000007266 UP000002358 UP000000311 UP000242457 UP000053105 UP000215335 UP000005205 UP000053825 UP000078541 UP000075809 UP000078492 UP000192223 UP000036403 UP000030742 UP000019118 UP000053097 UP000279307 UP000027135 UP000198287 UP000000305 UP000076858 UP000076502 UP000078542 UP000008237 UP000235965 UP000094527 UP000005408 UP000265140 UP000001357 UP000000437 UP000085678 UP000087266 UP000193380 UP000261540 UP000192224 UP000271974 UP000007755 UP000221080 UP000050525 UP000001554 UP000037510 UP000265160 UP000265100
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
H9J1E8
A0A2A4K9J9
A0A2H1VFD5
A0A194RJQ3
A0A194PKP4
A0A3S2LYP7
+ More
S4PHC9 I4DK39 A0A1E1WHA9 A0A212EXS6 A0A2P8YNA7 D6WT45 A0A0T6BB72 V5I9J7 K7J4Z1 A0A1Y1KTU2 J3JTW0 E1ZWG1 A0A2A3EMI3 V9II29 A0A0N0U515 A0A411G642 A0A232EYU2 A0A158NPP5 A0A0C9RFI6 A0A0L7R042 A0A195ESV4 A0A151X8H8 A0A195EM54 A0A1W4XAA1 A0A0J7L0J6 U4TYK1 N6TTR8 A0A026WTY8 A0A067R651 A0A226ERV9 U4U3C1 E9GZ78 A0A131XNR3 A0A224YG33 A0A131Z2F3 A0A023G7V6 A0A023FQ41 G3MRX7 A0A023FXX9 A0A0P5VFR9 A0A1E1X2R1 A0A0P6JVR0 A0A131XZ20 A0A154PEJ0 A0A0P5AUD2 A0A0P5WMP8 A0A0P4YC36 A0A0P5YL84 A0A0P5Y9B9 A0A151I8C8 E2BIK2 A0A2J7RR21 A0A023FNY4 A0A2R5LFX9 A0A1D2MXK3 A0A0P5BEP2 A0A0P4YZ09 K1QBT2 A0A3P9ADQ9 A9UWM7 A0A1Z5KYH6 F1RCM3 A0A1S3JWY0 B5X963 B5X8Y8 A0A060XEY9 A0A3B3RT78 C1BW52 A0A1W4Y7R5 A0A433UB52 A0A0P5RDI7 A0A3Q8QF21 B2GRB2 Q568V4 F4W7J5 B5XD66 O57332 R4UMP0 A0A444USC0 A0A060YSB5 A0A0P5ND39 A0A0K8RGM3 A0A1S3RIW7 A0A0B6ZXC6 A0A0P5N9Y1 W5USF9 A0A2D0QBT5 A0A151NTL8 A7RA95 C3ZIC3 A0A1P8DB30 A0A0L7KS96 A0A3P9DRT1 A0A3P8P4W9
S4PHC9 I4DK39 A0A1E1WHA9 A0A212EXS6 A0A2P8YNA7 D6WT45 A0A0T6BB72 V5I9J7 K7J4Z1 A0A1Y1KTU2 J3JTW0 E1ZWG1 A0A2A3EMI3 V9II29 A0A0N0U515 A0A411G642 A0A232EYU2 A0A158NPP5 A0A0C9RFI6 A0A0L7R042 A0A195ESV4 A0A151X8H8 A0A195EM54 A0A1W4XAA1 A0A0J7L0J6 U4TYK1 N6TTR8 A0A026WTY8 A0A067R651 A0A226ERV9 U4U3C1 E9GZ78 A0A131XNR3 A0A224YG33 A0A131Z2F3 A0A023G7V6 A0A023FQ41 G3MRX7 A0A023FXX9 A0A0P5VFR9 A0A1E1X2R1 A0A0P6JVR0 A0A131XZ20 A0A154PEJ0 A0A0P5AUD2 A0A0P5WMP8 A0A0P4YC36 A0A0P5YL84 A0A0P5Y9B9 A0A151I8C8 E2BIK2 A0A2J7RR21 A0A023FNY4 A0A2R5LFX9 A0A1D2MXK3 A0A0P5BEP2 A0A0P4YZ09 K1QBT2 A0A3P9ADQ9 A9UWM7 A0A1Z5KYH6 F1RCM3 A0A1S3JWY0 B5X963 B5X8Y8 A0A060XEY9 A0A3B3RT78 C1BW52 A0A1W4Y7R5 A0A433UB52 A0A0P5RDI7 A0A3Q8QF21 B2GRB2 Q568V4 F4W7J5 B5XD66 O57332 R4UMP0 A0A444USC0 A0A060YSB5 A0A0P5ND39 A0A0K8RGM3 A0A1S3RIW7 A0A0B6ZXC6 A0A0P5N9Y1 W5USF9 A0A2D0QBT5 A0A151NTL8 A7RA95 C3ZIC3 A0A1P8DB30 A0A0L7KS96 A0A3P9DRT1 A0A3P8P4W9
PDB
5M32
E-value=2.48668e-73,
Score=697
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
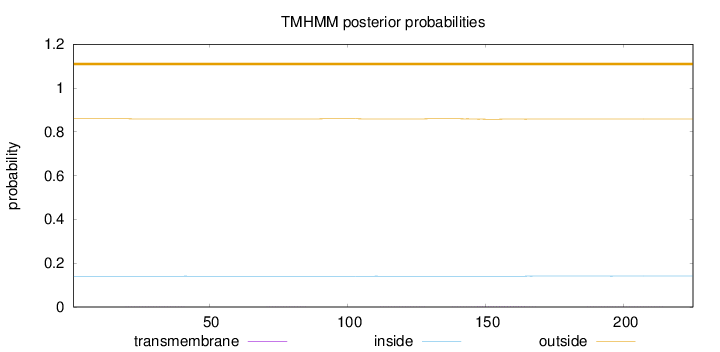
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05766
Exp number, first 60 AAs:
0.00855
Total prob of N-in:
0.14058
outside
1 - 225
Population Genetic Test Statistics
Pi
335.763737
Theta
193.338319
Tajima's D
2.456193
CLR
0
CSRT
0.941902904854757
Interpretation
Uncertain
