Pre Gene Modal
BGIBMGA003336
Annotation
PREDICTED:_mRNA_cap_guanine-N7_methyltransferase_[Papilio_machaon]
Full name
mRNA cap guanine-N7 methyltransferase
Alternative Name
RG7MT1
mRNA (guanine-N(7)-)-methyltransferase
mRNA cap methyltransferase
mRNA (guanine-N(7)-)-methyltransferase
mRNA cap methyltransferase
Location in the cell
Cytoplasmic Reliability : 1.968 Nuclear Reliability : 2.161
Sequence
CDS
ATGTCAGAATTGGGAGATTCCGAACTTTCGCTACAAAAACAAACTGAAAATGTCGATGAAAACAAAATCAATGCCAATAATTCAAATCATGAACCTCCTCCCAAAAAACATAGCAGCGTTGTAGCAGCACACTACAACCATCTGGAAGAAAAAGGTCTAAAAGAACGATACAATTCCCCAATATTTCACTTAAGGAATTTCAACAACTGGGTGAAAAGTGTTCTAATCCAAGAATTCACAGATAAAATACGCGAAAAAGACTACGGCCGTCCGCTCCGTATTTTAGACATCTGCTGTGGAAAAGGTGGAGATCTAAGTAAATGGCAAAAAGTTCGGCCGGAACATGCAATTTTCGCTGACATAGCTGAGGTTTCCGTTCAGCAATGCGAAACTCGTTACGAAGAACTTCGTAGGAGATGCGGTCGGCTCTTTTCAGCCGAATTCATTGCTACGGATTGTACAAAAGACACACTTAGAGATAAATACAAAGATCCATCAATCAGTTTTGATATAGTTAGCTGTCAATTTGGATTACATTATAGTTTTGAAAGCTTACCACAAGCAAGAAGAATGTTGACCAACATTTCTGAATGTCTAAAGCCTGGGGGCTATTTTTTTGGCACAATTCCGAATGCTAACGAGATTGTTTCAAGGGCACAGAAGACCCAAGATTACGCATTTGGTAACAGGATTTACAATATAAAACTACTGTTTGATCCCAAAGATGGCTATCCGCTTTTTGGAGCGAAATATGACTTTCACTTGGAAGGTGTGGTAGACTGTCCGGAGTTCTTAGTCAACTTTGAGCTGTTTAAGAAATTAGCTTCAGAATATGGCTTGGAATTAGTTTATAAAGCAGGCTTTGATCATTTTTATAAGGACCACTGGGAAAATCACAAACAGCTGTTTCAAAGGATAATGTGTTTTGAGACTTATCCAGCGCCAAATGGTAAAACCCTTATGGGTGAAGAGTCTGAATATGAGCACGCAAAAAAATACTGGGACAGTAAAGAGTATAAGGCTGACCAAAATTGTATCGGCACCATGAGTATATGTGAATGGGAGGTTGCTACAATATACATGGCCTTTGCATTTAAGAAATTGAAATGCACTCAGTGGAGTTCTGATGGGAAGCCAACTTATACAGTTTCCAAAGAAACTAATCAGTCAACTAGTAAGCAATGA
Protein
MSELGDSELSLQKQTENVDENKINANNSNHEPPPKKHSSVVAAHYNHLEEKGLKERYNSPIFHLRNFNNWVKSVLIQEFTDKIREKDYGRPLRILDICCGKGGDLSKWQKVRPEHAIFADIAEVSVQQCETRYEELRRRCGRLFSAEFIATDCTKDTLRDKYKDPSISFDIVSCQFGLHYSFESLPQARRMLTNISECLKPGGYFFGTIPNANEIVSRAQKTQDYAFGNRIYNIKLLFDPKDGYPLFGAKYDFHLEGVVDCPEFLVNFELFKKLASEYGLELVYKAGFDHFYKDHWENHKQLFQRIMCFETYPAPNGKTLMGEESEYEHAKKYWDSKEYKADQNCIGTMSICEWEVATIYMAFAFKKLKCTQWSSDGKPTYTVSKETNQSTSKQ
Summary
Description
Catalytic subunit of the mRNA-capping methyltransferase RNMT:RAMAC complex that methylates the N7 position of the added guanosine to the 5'-cap structure of mRNAs. Binds RNA containing 5'-terminal GpppC.
Catalytic subunit of the mRNA-capping methyltransferase RNMT:RAMAC complex that methylates the N7 position of the added guanosine to the 5'-cap structure of mRNAs (PubMed:9790902, PubMed:9705270, PubMed:10347220, PubMed:11114884, PubMed:22099306, PubMed:27422871). Binds RNA containing 5'-terminal GpppC (PubMed:11114884).
Catalytic subunit of the mRNA-capping methyltransferase RNMT:RAMAC complex that methylates the N7 position of the added guanosine to the 5'-cap structure of mRNAs (PubMed:9790902, PubMed:9705270, PubMed:10347220, PubMed:11114884, PubMed:22099306, PubMed:27422871). Binds RNA containing 5'-terminal GpppC (PubMed:11114884).
Catalytic Activity
a 5'-(5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(purine-ribonucleotide)-[mRNA] + S-adenosyl-L-homocysteine
Subunit
Interacts with importin alpha, leading to stimulate both RNA-binding and methyltransferase activity. Interaction with importin alpha and beta is required for its nuclear localization, importin beta dissociating in response to RanGTP, allowing RNMT-importin alpha to bind RNA substrates. Interacts with elongating form of polymerase II and RNGTT. Interacts with RAMAC, this interaction significantly enhances RNA-binding and cap methyltransferase activity.
Interacts with importin alpha, leading to stimulate both RNA-binding and methyltransferase activity (PubMed:11114884). Interaction with importin alpha and beta is required for its nuclear localization, importin beta dissociating in response to RanGTP, allowing RNMT-importin alpha to bind RNA substrates (PubMed:11114884). Interacts with elongating form of polymerase II and RNGTT (PubMed:9705270). Interacts with RAMAC, this interaction significantly enhances RNA-binding and cap methyltransferase activity (PubMed:22099306, Ref.14).
Interacts with importin alpha, leading to stimulate both RNA-binding and methyltransferase activity (PubMed:11114884). Interaction with importin alpha and beta is required for its nuclear localization, importin beta dissociating in response to RanGTP, allowing RNMT-importin alpha to bind RNA substrates (PubMed:11114884). Interacts with elongating form of polymerase II and RNGTT (PubMed:9705270). Interacts with RAMAC, this interaction significantly enhances RNA-binding and cap methyltransferase activity (PubMed:22099306, Ref.14).
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. mRNA cap 0 methyltransferase family.
Keywords
Complete proteome
Methyltransferase
mRNA capping
mRNA processing
Nucleus
Phosphoprotein
RNA-binding
S-adenosyl-L-methionine
Transferase
3D-structure
Alternative splicing
Reference proteome
Feature
chain mRNA cap guanine-N7 methyltransferase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J1E9
A0A437BEG1
A0A2A4J3S1
A0A2H1WTN5
A0A0N1IGT8
A0A0L7KQK7
+ More
A0A194PNY4 A0A212EJA2 A0A1B6G2R1 A0A1B0GGR5 A0A232F6X0 A0A2J7Q1K7 A0A1L8DGS0 K7J100 A0A0P4VGV1 A0A1L8DGV5 E2BJP0 A0A0T6AUK7 A0A224XQY9 A0A0V0G781 A0A0K8TQX8 A0A182GML6 A0A1B6DGR4 A0A1B6MB57 E2AAT3 A0A1S4FUB6 A0A023F1P1 A0A026X3T5 Q16PB9 A0A0C9RMK3 A0A069DYP2 A0A158NS92 F4W6S2 A0A195CWU6 A0A1B0FY63 A0A067RLV0 A0A1Q3F4U5 A0A1Q3F553 A0A1B3PDG3 B0WEZ1 A0A151J8A6 D6WKN1 V5GCT7 A0A2J7Q1L5 A0A2P1L4C9 A0A1B3PDB0 A0A2A3EH70 A0A433UAH5 A0A0L7QVX7 A0A088A1M2 A0A210R621 U5ERD1 A0A151XK11 A0A2Y9LP65 A0A1S3G1R1 G7PWB3 A0A2U4A5N0 A0A2Y9FBG7 A0A384B9U3 A0A341CXR4 A0A1W4XGK1 A0A2K5LQU3 A0A2C9JTM7 A0A340XLD8 A0A0D9RY29 A0A096N0L4 G7NK78 Q4R7K1 A0A2K6QXR6 G1RDH3 A0A2K6TUM3 A0A452QEC1 A0A384DMH7 A0A2K5Z2W8 A0A2K5E337 U3EIZ4 A0A0L0C1H0 A0A2K5IUE0 F1MHQ5 A6H761 O43148 H2R6V8 A0A2R8ZPU1 G3QRZ1 M3XYX8 A0A3Q1M102 A0A0K8VIP5 A0A3Q7VGV8 G3TZ46 A0A1S3A4J7 A0A452DL89 W5NVX3 I3M5X4 A0A0A9YUQ9 G1MH91 L8IQE5 A0A146L245 N6TRS5 A0A091GJR7
A0A194PNY4 A0A212EJA2 A0A1B6G2R1 A0A1B0GGR5 A0A232F6X0 A0A2J7Q1K7 A0A1L8DGS0 K7J100 A0A0P4VGV1 A0A1L8DGV5 E2BJP0 A0A0T6AUK7 A0A224XQY9 A0A0V0G781 A0A0K8TQX8 A0A182GML6 A0A1B6DGR4 A0A1B6MB57 E2AAT3 A0A1S4FUB6 A0A023F1P1 A0A026X3T5 Q16PB9 A0A0C9RMK3 A0A069DYP2 A0A158NS92 F4W6S2 A0A195CWU6 A0A1B0FY63 A0A067RLV0 A0A1Q3F4U5 A0A1Q3F553 A0A1B3PDG3 B0WEZ1 A0A151J8A6 D6WKN1 V5GCT7 A0A2J7Q1L5 A0A2P1L4C9 A0A1B3PDB0 A0A2A3EH70 A0A433UAH5 A0A0L7QVX7 A0A088A1M2 A0A210R621 U5ERD1 A0A151XK11 A0A2Y9LP65 A0A1S3G1R1 G7PWB3 A0A2U4A5N0 A0A2Y9FBG7 A0A384B9U3 A0A341CXR4 A0A1W4XGK1 A0A2K5LQU3 A0A2C9JTM7 A0A340XLD8 A0A0D9RY29 A0A096N0L4 G7NK78 Q4R7K1 A0A2K6QXR6 G1RDH3 A0A2K6TUM3 A0A452QEC1 A0A384DMH7 A0A2K5Z2W8 A0A2K5E337 U3EIZ4 A0A0L0C1H0 A0A2K5IUE0 F1MHQ5 A6H761 O43148 H2R6V8 A0A2R8ZPU1 G3QRZ1 M3XYX8 A0A3Q1M102 A0A0K8VIP5 A0A3Q7VGV8 G3TZ46 A0A1S3A4J7 A0A452DL89 W5NVX3 I3M5X4 A0A0A9YUQ9 G1MH91 L8IQE5 A0A146L245 N6TRS5 A0A091GJR7
EC Number
2.1.1.56
Pubmed
19121390
26354079
26227816
22118469
28648823
20075255
+ More
27129103 20798317 26369729 26483478 25474469 24508170 30249741 17510324 26334808 21347285 21719571 24845553 18362917 19820115 29501422 28812685 22002653 15562597 17431167 25319552 25362486 24813606 25243066 26108605 19393038 9790902 9705270 10589710 9455477 15489334 10347220 11114884 15767670 21269460 22099306 27422871 16136131 22722832 22398555 20809919 25401762 20010809 22751099 26823975 23537049
27129103 20798317 26369729 26483478 25474469 24508170 30249741 17510324 26334808 21347285 21719571 24845553 18362917 19820115 29501422 28812685 22002653 15562597 17431167 25319552 25362486 24813606 25243066 26108605 19393038 9790902 9705270 10589710 9455477 15489334 10347220 11114884 15767670 21269460 22099306 27422871 16136131 22722832 22398555 20809919 25401762 20010809 22751099 26823975 23537049
EMBL
BABH01023826
RSAL01000077
RVE48809.1
NWSH01003201
PCG66797.1
ODYU01010974
+ More
SOQ56418.1 KQ460709 KPJ12716.1 JTDY01006928 KOB65588.1 KQ459598 KPI94693.1 AGBW02014506 OWR41551.1 GECZ01013040 JAS56729.1 AJWK01000610 NNAY01000821 OXU26332.1 NEVH01019395 PNF22472.1 GFDF01008432 JAV05652.1 GDKW01003025 JAI53570.1 GFDF01008467 JAV05617.1 GL448604 EFN84098.1 LJIG01022805 KRT78621.1 GFTR01005566 JAW10860.1 GECL01002834 JAP03290.1 GDAI01000836 JAI16767.1 JXUM01001747 KQ560127 KXJ84322.1 GEDC01012419 JAS24879.1 GEBQ01006812 JAT33165.1 GL438172 EFN69462.1 GBBI01003489 JAC15223.1 KK107019 QOIP01000007 EZA62748.1 RLU20528.1 CH477788 EAT36219.1 GBYB01008211 JAG77978.1 GBGD01002045 JAC86844.1 ADTU01024723 GL887726 EGI70084.1 KQ977171 KYN05141.1 AJVK01031282 KK852432 KDR24008.1 GFDL01012465 JAV22580.1 GFDL01012412 JAV22633.1 KU659970 AOG17768.1 DS231912 EDS25849.1 KQ979602 KYN20525.1 KQ971342 EFA03004.2 GALX01000526 JAB67940.1 PNF22471.1 MG596905 AVP12660.1 KU659917 AOG17716.1 KZ288250 PBC31077.1 RQTK01000024 RUS90792.1 KQ414722 KOC62706.1 NEDP02000186 OWF56493.1 GANO01002856 JAB57015.1 KQ982045 KYQ60744.1 CM001293 EHH58736.1 AQIB01129447 AHZZ02012178 AHZZ02012179 JSUE03019118 JSUE03019119 JU336902 JU477644 JV048578 CM001270 AFE80655.1 AFH34448.1 AFI38649.1 EHH29151.1 AB168817 ADFV01096840 GAMT01001012 GAMS01002790 GAMR01008598 GAMQ01004106 GAMP01008253 JAB10849.1 JAB20346.1 JAB25334.1 JAB37745.1 JAB44502.1 JRES01001003 KNC26203.1 BC146125 AAI46126.1 AB022604 AB022605 AF067791 AB020966 AB007858 EF445026 CH471113 BC036798 AACZ04060022 GABC01008892 GABF01003328 GABD01003664 GABE01008602 NBAG03000406 JAA02446.1 JAA18817.1 JAA29436.1 JAA36137.1 PNI28336.1 AJFE02023052 CABD030106927 AEYP01011347 AEYP01011348 GDHF01013587 JAI38727.1 LWLT01000023 AMGL01066958 AGTP01079821 GBHO01007675 GBRD01000703 JAG35929.1 JAG65118.1 ACTA01067257 ACTA01075257 ACTA01083257 ACTA01091257 ACTA01099257 JH880802 ELR58800.1 GDHC01016620 JAQ02009.1 APGK01054294 APGK01054295 APGK01054296 KB741248 KB631794 ENN71965.1 ERL86187.1 KL448232 KFO81419.1
SOQ56418.1 KQ460709 KPJ12716.1 JTDY01006928 KOB65588.1 KQ459598 KPI94693.1 AGBW02014506 OWR41551.1 GECZ01013040 JAS56729.1 AJWK01000610 NNAY01000821 OXU26332.1 NEVH01019395 PNF22472.1 GFDF01008432 JAV05652.1 GDKW01003025 JAI53570.1 GFDF01008467 JAV05617.1 GL448604 EFN84098.1 LJIG01022805 KRT78621.1 GFTR01005566 JAW10860.1 GECL01002834 JAP03290.1 GDAI01000836 JAI16767.1 JXUM01001747 KQ560127 KXJ84322.1 GEDC01012419 JAS24879.1 GEBQ01006812 JAT33165.1 GL438172 EFN69462.1 GBBI01003489 JAC15223.1 KK107019 QOIP01000007 EZA62748.1 RLU20528.1 CH477788 EAT36219.1 GBYB01008211 JAG77978.1 GBGD01002045 JAC86844.1 ADTU01024723 GL887726 EGI70084.1 KQ977171 KYN05141.1 AJVK01031282 KK852432 KDR24008.1 GFDL01012465 JAV22580.1 GFDL01012412 JAV22633.1 KU659970 AOG17768.1 DS231912 EDS25849.1 KQ979602 KYN20525.1 KQ971342 EFA03004.2 GALX01000526 JAB67940.1 PNF22471.1 MG596905 AVP12660.1 KU659917 AOG17716.1 KZ288250 PBC31077.1 RQTK01000024 RUS90792.1 KQ414722 KOC62706.1 NEDP02000186 OWF56493.1 GANO01002856 JAB57015.1 KQ982045 KYQ60744.1 CM001293 EHH58736.1 AQIB01129447 AHZZ02012178 AHZZ02012179 JSUE03019118 JSUE03019119 JU336902 JU477644 JV048578 CM001270 AFE80655.1 AFH34448.1 AFI38649.1 EHH29151.1 AB168817 ADFV01096840 GAMT01001012 GAMS01002790 GAMR01008598 GAMQ01004106 GAMP01008253 JAB10849.1 JAB20346.1 JAB25334.1 JAB37745.1 JAB44502.1 JRES01001003 KNC26203.1 BC146125 AAI46126.1 AB022604 AB022605 AF067791 AB020966 AB007858 EF445026 CH471113 BC036798 AACZ04060022 GABC01008892 GABF01003328 GABD01003664 GABE01008602 NBAG03000406 JAA02446.1 JAA18817.1 JAA29436.1 JAA36137.1 PNI28336.1 AJFE02023052 CABD030106927 AEYP01011347 AEYP01011348 GDHF01013587 JAI38727.1 LWLT01000023 AMGL01066958 AGTP01079821 GBHO01007675 GBRD01000703 JAG35929.1 JAG65118.1 ACTA01067257 ACTA01075257 ACTA01083257 ACTA01091257 ACTA01099257 JH880802 ELR58800.1 GDHC01016620 JAQ02009.1 APGK01054294 APGK01054295 APGK01054296 KB741248 KB631794 ENN71965.1 ERL86187.1 KL448232 KFO81419.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000037510
UP000053268
+ More
UP000007151 UP000092461 UP000215335 UP000235965 UP000002358 UP000008237 UP000069940 UP000249989 UP000000311 UP000053097 UP000279307 UP000008820 UP000005205 UP000007755 UP000078542 UP000092462 UP000027135 UP000002320 UP000078492 UP000007266 UP000242457 UP000271974 UP000053825 UP000005203 UP000242188 UP000075809 UP000248483 UP000081671 UP000009130 UP000245320 UP000248484 UP000261681 UP000252040 UP000192223 UP000233060 UP000076420 UP000265300 UP000029965 UP000028761 UP000006718 UP000233100 UP000233200 UP000001073 UP000233220 UP000291022 UP000261680 UP000291021 UP000233140 UP000233020 UP000008225 UP000037069 UP000233080 UP000009136 UP000005640 UP000002277 UP000240080 UP000001519 UP000000715 UP000286642 UP000007646 UP000079721 UP000291000 UP000002356 UP000005215 UP000008912 UP000019118 UP000030742 UP000053760
UP000007151 UP000092461 UP000215335 UP000235965 UP000002358 UP000008237 UP000069940 UP000249989 UP000000311 UP000053097 UP000279307 UP000008820 UP000005205 UP000007755 UP000078542 UP000092462 UP000027135 UP000002320 UP000078492 UP000007266 UP000242457 UP000271974 UP000053825 UP000005203 UP000242188 UP000075809 UP000248483 UP000081671 UP000009130 UP000245320 UP000248484 UP000261681 UP000252040 UP000192223 UP000233060 UP000076420 UP000265300 UP000029965 UP000028761 UP000006718 UP000233100 UP000233200 UP000001073 UP000233220 UP000291022 UP000261680 UP000291021 UP000233140 UP000233020 UP000008225 UP000037069 UP000233080 UP000009136 UP000005640 UP000002277 UP000240080 UP000001519 UP000000715 UP000286642 UP000007646 UP000079721 UP000291000 UP000002356 UP000005215 UP000008912 UP000019118 UP000030742 UP000053760
Pfam
PF03291 Pox_MCEL
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J1E9
A0A437BEG1
A0A2A4J3S1
A0A2H1WTN5
A0A0N1IGT8
A0A0L7KQK7
+ More
A0A194PNY4 A0A212EJA2 A0A1B6G2R1 A0A1B0GGR5 A0A232F6X0 A0A2J7Q1K7 A0A1L8DGS0 K7J100 A0A0P4VGV1 A0A1L8DGV5 E2BJP0 A0A0T6AUK7 A0A224XQY9 A0A0V0G781 A0A0K8TQX8 A0A182GML6 A0A1B6DGR4 A0A1B6MB57 E2AAT3 A0A1S4FUB6 A0A023F1P1 A0A026X3T5 Q16PB9 A0A0C9RMK3 A0A069DYP2 A0A158NS92 F4W6S2 A0A195CWU6 A0A1B0FY63 A0A067RLV0 A0A1Q3F4U5 A0A1Q3F553 A0A1B3PDG3 B0WEZ1 A0A151J8A6 D6WKN1 V5GCT7 A0A2J7Q1L5 A0A2P1L4C9 A0A1B3PDB0 A0A2A3EH70 A0A433UAH5 A0A0L7QVX7 A0A088A1M2 A0A210R621 U5ERD1 A0A151XK11 A0A2Y9LP65 A0A1S3G1R1 G7PWB3 A0A2U4A5N0 A0A2Y9FBG7 A0A384B9U3 A0A341CXR4 A0A1W4XGK1 A0A2K5LQU3 A0A2C9JTM7 A0A340XLD8 A0A0D9RY29 A0A096N0L4 G7NK78 Q4R7K1 A0A2K6QXR6 G1RDH3 A0A2K6TUM3 A0A452QEC1 A0A384DMH7 A0A2K5Z2W8 A0A2K5E337 U3EIZ4 A0A0L0C1H0 A0A2K5IUE0 F1MHQ5 A6H761 O43148 H2R6V8 A0A2R8ZPU1 G3QRZ1 M3XYX8 A0A3Q1M102 A0A0K8VIP5 A0A3Q7VGV8 G3TZ46 A0A1S3A4J7 A0A452DL89 W5NVX3 I3M5X4 A0A0A9YUQ9 G1MH91 L8IQE5 A0A146L245 N6TRS5 A0A091GJR7
A0A194PNY4 A0A212EJA2 A0A1B6G2R1 A0A1B0GGR5 A0A232F6X0 A0A2J7Q1K7 A0A1L8DGS0 K7J100 A0A0P4VGV1 A0A1L8DGV5 E2BJP0 A0A0T6AUK7 A0A224XQY9 A0A0V0G781 A0A0K8TQX8 A0A182GML6 A0A1B6DGR4 A0A1B6MB57 E2AAT3 A0A1S4FUB6 A0A023F1P1 A0A026X3T5 Q16PB9 A0A0C9RMK3 A0A069DYP2 A0A158NS92 F4W6S2 A0A195CWU6 A0A1B0FY63 A0A067RLV0 A0A1Q3F4U5 A0A1Q3F553 A0A1B3PDG3 B0WEZ1 A0A151J8A6 D6WKN1 V5GCT7 A0A2J7Q1L5 A0A2P1L4C9 A0A1B3PDB0 A0A2A3EH70 A0A433UAH5 A0A0L7QVX7 A0A088A1M2 A0A210R621 U5ERD1 A0A151XK11 A0A2Y9LP65 A0A1S3G1R1 G7PWB3 A0A2U4A5N0 A0A2Y9FBG7 A0A384B9U3 A0A341CXR4 A0A1W4XGK1 A0A2K5LQU3 A0A2C9JTM7 A0A340XLD8 A0A0D9RY29 A0A096N0L4 G7NK78 Q4R7K1 A0A2K6QXR6 G1RDH3 A0A2K6TUM3 A0A452QEC1 A0A384DMH7 A0A2K5Z2W8 A0A2K5E337 U3EIZ4 A0A0L0C1H0 A0A2K5IUE0 F1MHQ5 A6H761 O43148 H2R6V8 A0A2R8ZPU1 G3QRZ1 M3XYX8 A0A3Q1M102 A0A0K8VIP5 A0A3Q7VGV8 G3TZ46 A0A1S3A4J7 A0A452DL89 W5NVX3 I3M5X4 A0A0A9YUQ9 G1MH91 L8IQE5 A0A146L245 N6TRS5 A0A091GJR7
PDB
3EPP
E-value=6.16596e-80,
Score=757
Ontologies
GO
PANTHER
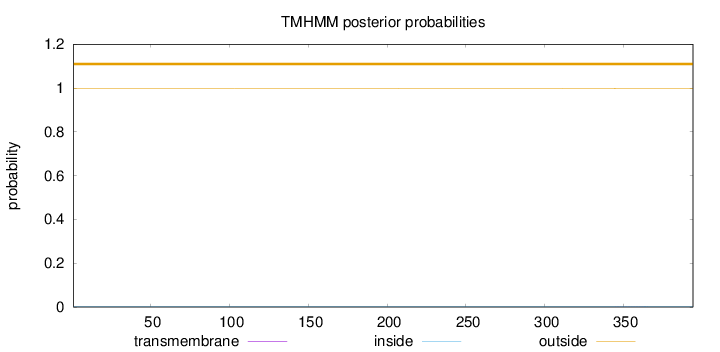
Topology
Subcellular location
Nucleus
Length:
394
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0205
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00164
outside
1 - 394
Population Genetic Test Statistics
Pi
200.861197
Theta
173.869344
Tajima's D
0.472677
CLR
0.135745
CSRT
0.504474776261187
Interpretation
Uncertain
