Pre Gene Modal
BGIBMGA003338
Annotation
mitochondrial_28S_ribosomal_protein_S14_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.974
Sequence
CDS
ATGAACTTAAATATCTCAATTTTAGGAAAATTCATTCTAAAATCACAATCAATAGCCGGAGTCGGGTATCAACAAGTACGGAATAAATGGGCTAACTGGCTCATGATTCGTGATGTAAAGCGACGTAGATTGAGTGCAGAGAACTTTCTTGAAAGGACCAGAGTTAATGCATTGCGGAAAAATGACATCCTCCCTGTTGAAATCAGAGAGATAGCTGACAAGCAAATCAATCAGTTTGAATGGAATGCCACACCTATGAGAATTAACAACAGATGTGTTATCACTTCACGACCCAGAGGTATAGTAAAGGAATGGCGCATGAGTCGTATAGTCTGGAGGCATCTAGCGGATTACAACAAACTCTCAGGAGTACAGAGGGCTATGTGGGGCTGA
Protein
MNLNISILGKFILKSQSIAGVGYQQVRNKWANWLMIRDVKRRRLSAENFLERTRVNALRKNDILPVEIREIADKQINQFEWNATPMRINNRCVITSRPRGIVKEWRMSRIVWRHLADYNKLSGVQRAMWG
Summary
Similarity
Belongs to the carnitine/choline acetyltransferase family.
Uniprot
Q1HPY6
A0A3S2L6Y2
A0A212EXX5
A0A2A4KB43
A0A194RIX5
A0A0L7KUT0
+ More
A0A2H1VXC1 H9J1F1 A0A1I8MEB8 A0A0M3QZG6 B4MA46 B4L6X2 B4JX26 A0JQ38 Q9VWI3 B4PZI4 B4I750 B3NVN3 A0A1W4VST0 W8BVV5 B3MPT6 A0A0L0C403 B5M0Y1 A0A1I8P2T0 B4NPI7 B5DL77 B4GW64 A0A1A9WXX4 A0A084VBZ6 A0A0K8UUE0 A0A023EE59 A0A1L8EH77 A0A3B0KVU7 A0A067R2A8 A0A1B0BK15 A0A1B0G1J0 T1EA22 A0A2M3ZEW7 A0A2P8XSK7 Q17DU5 A0A182FG85 A0A182LUP5 A0A2M4C347 A0A2M4AY10 A0A2M4AYA9 A0A2M4C3R1 W5J7X8 A0A182R9S4 A0A2M4C371 D6WKQ4 A0A182YFH0 A0A182UR13 A0A1Q3FCV1 A0A2C9GRW0 A0A182UCX5 A0A182KUS5 Q7PW63 A0A182X2Y7 A0A182PDF6 A0A182SFX0 A0A182K3P8 A0A2J7QL97 A0A182J4I6 B0WSJ3 A0A1A9XRZ8 A0A1Y9H9K2 A0A023EFL4 A0A182WGW9 A0A3B3QP36 F6YV30 B3N1J1 Q28D63 A0A0E9WHN3 A0A0M3KL53 A0A1L8GG11 A0A1Y1M9A5 A0A340WWG8 A0A2Y9TIM0 A0A336MQH8 A0A1B0CLF4 A0A287D5W5 A0A1S3WTS4 A0A452IPJ9 A0A401RJS9 A0A0K8TLA2 F7C1X2 A0A341CKS4 A0A2U3V8J5 A0A2Y9LRA9 A0A0Q3X046 H0WWY2 F6SVT1 A0A1U7RP09 U3K160 A0A0A9X4R2 A0A383YRE3 A0A1W5A6I2 G5ARS1 A0A2Y9DEG4
A0A2H1VXC1 H9J1F1 A0A1I8MEB8 A0A0M3QZG6 B4MA46 B4L6X2 B4JX26 A0JQ38 Q9VWI3 B4PZI4 B4I750 B3NVN3 A0A1W4VST0 W8BVV5 B3MPT6 A0A0L0C403 B5M0Y1 A0A1I8P2T0 B4NPI7 B5DL77 B4GW64 A0A1A9WXX4 A0A084VBZ6 A0A0K8UUE0 A0A023EE59 A0A1L8EH77 A0A3B0KVU7 A0A067R2A8 A0A1B0BK15 A0A1B0G1J0 T1EA22 A0A2M3ZEW7 A0A2P8XSK7 Q17DU5 A0A182FG85 A0A182LUP5 A0A2M4C347 A0A2M4AY10 A0A2M4AYA9 A0A2M4C3R1 W5J7X8 A0A182R9S4 A0A2M4C371 D6WKQ4 A0A182YFH0 A0A182UR13 A0A1Q3FCV1 A0A2C9GRW0 A0A182UCX5 A0A182KUS5 Q7PW63 A0A182X2Y7 A0A182PDF6 A0A182SFX0 A0A182K3P8 A0A2J7QL97 A0A182J4I6 B0WSJ3 A0A1A9XRZ8 A0A1Y9H9K2 A0A023EFL4 A0A182WGW9 A0A3B3QP36 F6YV30 B3N1J1 Q28D63 A0A0E9WHN3 A0A0M3KL53 A0A1L8GG11 A0A1Y1M9A5 A0A340WWG8 A0A2Y9TIM0 A0A336MQH8 A0A1B0CLF4 A0A287D5W5 A0A1S3WTS4 A0A452IPJ9 A0A401RJS9 A0A0K8TLA2 F7C1X2 A0A341CKS4 A0A2U3V8J5 A0A2Y9LRA9 A0A0Q3X046 H0WWY2 F6SVT1 A0A1U7RP09 U3K160 A0A0A9X4R2 A0A383YRE3 A0A1W5A6I2 G5ARS1 A0A2Y9DEG4
Pubmed
22118469
26354079
26227816
19121390
25315136
17994087
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 26108605 19166301 15632085 24438588 24945155 26483478 24845553 29403074 17510324 20920257 23761445 18362917 19820115 25244985 20966253 12364791 14747013 17210077 29240929 20431018 25613341 25837512 30089917 30723633 27762356 28004739 28562605 30297745 26369729 19892987 17495919 25401762 26823975 21993625
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24495485 26108605 19166301 15632085 24438588 24945155 26483478 24845553 29403074 17510324 20920257 23761445 18362917 19820115 25244985 20966253 12364791 14747013 17210077 29240929 20431018 25613341 25837512 30089917 30723633 27762356 28004739 28562605 30297745 26369729 19892987 17495919 25401762 26823975 21993625
EMBL
DQ443266
ABF51355.1
RSAL01000112
RVE47036.1
AGBW02011690
OWR46294.1
+ More
NWSH01000012 PCG80882.1 KQ460106 KPJ17778.1 JTDY01005421 KOB67013.1 ODYU01005012 SOQ45480.1 BABH01023826 CP012528 ALC49339.1 CH940655 EDW66105.1 CH933812 EDW06118.1 CH916376 EDV95302.1 BT029408 ABK57065.1 AE014298 BT023293 KX532149 AAF48956.2 AAY55709.1 AHN59908.1 ANY27959.1 CM000162 EDX02140.1 CH480823 EDW56148.1 CH954180 EDV46698.1 GAMC01005502 JAC01054.1 CH902621 EDV44698.1 JRES01000940 KNC27010.1 EU930317 ACH56945.1 CH964291 EDW86427.1 CH379063 EDY72276.1 CH479193 EDW26909.1 ATLV01009878 KE524552 KFB35490.1 GDHF01022047 JAI30267.1 JXUM01053997 GAPW01005976 KQ561800 JAC07622.1 KXJ77487.1 GFDG01000785 JAV18014.1 OUUW01000044 SPP89996.1 KK852751 KDR17160.1 JXJN01015766 CCAG010002137 GAMD01001398 JAB00193.1 GGFM01006335 MBW27086.1 PYGN01001417 PSN34990.1 CH477291 EAT44567.1 AXCM01001875 GGFJ01010605 MBW59746.1 GGFK01012368 MBW45689.1 GGFK01012247 MBW45568.1 GGFJ01010670 MBW59811.1 ADMH02002078 ETN59493.1 GGFJ01010603 MBW59744.1 KQ971342 EFA04750.1 GFDL01009713 JAV25332.1 APCN01003737 AAAB01008984 EAA14742.3 NEVH01013250 PNF29362.1 DS232072 EDS33923.1 AXCN02002233 GAPW01005977 JAC07621.1 AAMC01000528 CH902658 EDV34917.2 CR855683 CAJ83488.1 GBXM01019579 JAH88998.1 AEMK02000070 DQIR01116143 HDA71619.1 CM004473 OCT82781.1 GEZM01037132 JAV82151.1 UFQT01002045 SSX32506.1 AJWK01017204 AGTP01014968 BEZZ01002941 GCC18395.1 GDAI01002476 JAI15127.1 LMAW01000377 KQL59260.1 AAQR03038574 AGTO01001091 AGTO01011210 AGTO01021330 GBHO01031529 GBRD01010968 GDHC01012924 JAG12075.1 JAG54856.1 JAQ05705.1 JH166641 GEBF01007353 EHA99731.1 JAN96279.1
NWSH01000012 PCG80882.1 KQ460106 KPJ17778.1 JTDY01005421 KOB67013.1 ODYU01005012 SOQ45480.1 BABH01023826 CP012528 ALC49339.1 CH940655 EDW66105.1 CH933812 EDW06118.1 CH916376 EDV95302.1 BT029408 ABK57065.1 AE014298 BT023293 KX532149 AAF48956.2 AAY55709.1 AHN59908.1 ANY27959.1 CM000162 EDX02140.1 CH480823 EDW56148.1 CH954180 EDV46698.1 GAMC01005502 JAC01054.1 CH902621 EDV44698.1 JRES01000940 KNC27010.1 EU930317 ACH56945.1 CH964291 EDW86427.1 CH379063 EDY72276.1 CH479193 EDW26909.1 ATLV01009878 KE524552 KFB35490.1 GDHF01022047 JAI30267.1 JXUM01053997 GAPW01005976 KQ561800 JAC07622.1 KXJ77487.1 GFDG01000785 JAV18014.1 OUUW01000044 SPP89996.1 KK852751 KDR17160.1 JXJN01015766 CCAG010002137 GAMD01001398 JAB00193.1 GGFM01006335 MBW27086.1 PYGN01001417 PSN34990.1 CH477291 EAT44567.1 AXCM01001875 GGFJ01010605 MBW59746.1 GGFK01012368 MBW45689.1 GGFK01012247 MBW45568.1 GGFJ01010670 MBW59811.1 ADMH02002078 ETN59493.1 GGFJ01010603 MBW59744.1 KQ971342 EFA04750.1 GFDL01009713 JAV25332.1 APCN01003737 AAAB01008984 EAA14742.3 NEVH01013250 PNF29362.1 DS232072 EDS33923.1 AXCN02002233 GAPW01005977 JAC07621.1 AAMC01000528 CH902658 EDV34917.2 CR855683 CAJ83488.1 GBXM01019579 JAH88998.1 AEMK02000070 DQIR01116143 HDA71619.1 CM004473 OCT82781.1 GEZM01037132 JAV82151.1 UFQT01002045 SSX32506.1 AJWK01017204 AGTP01014968 BEZZ01002941 GCC18395.1 GDAI01002476 JAI15127.1 LMAW01000377 KQL59260.1 AAQR03038574 AGTO01001091 AGTO01011210 AGTO01021330 GBHO01031529 GBRD01010968 GDHC01012924 JAG12075.1 JAG54856.1 JAQ05705.1 JH166641 GEBF01007353 EHA99731.1 JAN96279.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053240
UP000037510
UP000005204
+ More
UP000095301 UP000092553 UP000008792 UP000009192 UP000001070 UP000000803 UP000002282 UP000001292 UP000008711 UP000192221 UP000007801 UP000037069 UP000095300 UP000007798 UP000001819 UP000008744 UP000091820 UP000030765 UP000069940 UP000249989 UP000268350 UP000027135 UP000092460 UP000092444 UP000245037 UP000008820 UP000069272 UP000075883 UP000000673 UP000075900 UP000007266 UP000076408 UP000075903 UP000075840 UP000075902 UP000075882 UP000007062 UP000076407 UP000075885 UP000075901 UP000075881 UP000235965 UP000075880 UP000002320 UP000092443 UP000075886 UP000075920 UP000261540 UP000008143 UP000008227 UP000186698 UP000265300 UP000248484 UP000092461 UP000005215 UP000079721 UP000291020 UP000287033 UP000002281 UP000252040 UP000245320 UP000248483 UP000051836 UP000005225 UP000002280 UP000189705 UP000016665 UP000261681 UP000192224 UP000006813 UP000248480
UP000095301 UP000092553 UP000008792 UP000009192 UP000001070 UP000000803 UP000002282 UP000001292 UP000008711 UP000192221 UP000007801 UP000037069 UP000095300 UP000007798 UP000001819 UP000008744 UP000091820 UP000030765 UP000069940 UP000249989 UP000268350 UP000027135 UP000092460 UP000092444 UP000245037 UP000008820 UP000069272 UP000075883 UP000000673 UP000075900 UP000007266 UP000076408 UP000075903 UP000075840 UP000075902 UP000075882 UP000007062 UP000076407 UP000075885 UP000075901 UP000075881 UP000235965 UP000075880 UP000002320 UP000092443 UP000075886 UP000075920 UP000261540 UP000008143 UP000008227 UP000186698 UP000265300 UP000248484 UP000092461 UP000005215 UP000079721 UP000291020 UP000287033 UP000002281 UP000252040 UP000245320 UP000248483 UP000051836 UP000005225 UP000002280 UP000189705 UP000016665 UP000261681 UP000192224 UP000006813 UP000248480
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
ProteinModelPortal
Q1HPY6
A0A3S2L6Y2
A0A212EXX5
A0A2A4KB43
A0A194RIX5
A0A0L7KUT0
+ More
A0A2H1VXC1 H9J1F1 A0A1I8MEB8 A0A0M3QZG6 B4MA46 B4L6X2 B4JX26 A0JQ38 Q9VWI3 B4PZI4 B4I750 B3NVN3 A0A1W4VST0 W8BVV5 B3MPT6 A0A0L0C403 B5M0Y1 A0A1I8P2T0 B4NPI7 B5DL77 B4GW64 A0A1A9WXX4 A0A084VBZ6 A0A0K8UUE0 A0A023EE59 A0A1L8EH77 A0A3B0KVU7 A0A067R2A8 A0A1B0BK15 A0A1B0G1J0 T1EA22 A0A2M3ZEW7 A0A2P8XSK7 Q17DU5 A0A182FG85 A0A182LUP5 A0A2M4C347 A0A2M4AY10 A0A2M4AYA9 A0A2M4C3R1 W5J7X8 A0A182R9S4 A0A2M4C371 D6WKQ4 A0A182YFH0 A0A182UR13 A0A1Q3FCV1 A0A2C9GRW0 A0A182UCX5 A0A182KUS5 Q7PW63 A0A182X2Y7 A0A182PDF6 A0A182SFX0 A0A182K3P8 A0A2J7QL97 A0A182J4I6 B0WSJ3 A0A1A9XRZ8 A0A1Y9H9K2 A0A023EFL4 A0A182WGW9 A0A3B3QP36 F6YV30 B3N1J1 Q28D63 A0A0E9WHN3 A0A0M3KL53 A0A1L8GG11 A0A1Y1M9A5 A0A340WWG8 A0A2Y9TIM0 A0A336MQH8 A0A1B0CLF4 A0A287D5W5 A0A1S3WTS4 A0A452IPJ9 A0A401RJS9 A0A0K8TLA2 F7C1X2 A0A341CKS4 A0A2U3V8J5 A0A2Y9LRA9 A0A0Q3X046 H0WWY2 F6SVT1 A0A1U7RP09 U3K160 A0A0A9X4R2 A0A383YRE3 A0A1W5A6I2 G5ARS1 A0A2Y9DEG4
A0A2H1VXC1 H9J1F1 A0A1I8MEB8 A0A0M3QZG6 B4MA46 B4L6X2 B4JX26 A0JQ38 Q9VWI3 B4PZI4 B4I750 B3NVN3 A0A1W4VST0 W8BVV5 B3MPT6 A0A0L0C403 B5M0Y1 A0A1I8P2T0 B4NPI7 B5DL77 B4GW64 A0A1A9WXX4 A0A084VBZ6 A0A0K8UUE0 A0A023EE59 A0A1L8EH77 A0A3B0KVU7 A0A067R2A8 A0A1B0BK15 A0A1B0G1J0 T1EA22 A0A2M3ZEW7 A0A2P8XSK7 Q17DU5 A0A182FG85 A0A182LUP5 A0A2M4C347 A0A2M4AY10 A0A2M4AYA9 A0A2M4C3R1 W5J7X8 A0A182R9S4 A0A2M4C371 D6WKQ4 A0A182YFH0 A0A182UR13 A0A1Q3FCV1 A0A2C9GRW0 A0A182UCX5 A0A182KUS5 Q7PW63 A0A182X2Y7 A0A182PDF6 A0A182SFX0 A0A182K3P8 A0A2J7QL97 A0A182J4I6 B0WSJ3 A0A1A9XRZ8 A0A1Y9H9K2 A0A023EFL4 A0A182WGW9 A0A3B3QP36 F6YV30 B3N1J1 Q28D63 A0A0E9WHN3 A0A0M3KL53 A0A1L8GG11 A0A1Y1M9A5 A0A340WWG8 A0A2Y9TIM0 A0A336MQH8 A0A1B0CLF4 A0A287D5W5 A0A1S3WTS4 A0A452IPJ9 A0A401RJS9 A0A0K8TLA2 F7C1X2 A0A341CKS4 A0A2U3V8J5 A0A2Y9LRA9 A0A0Q3X046 H0WWY2 F6SVT1 A0A1U7RP09 U3K160 A0A0A9X4R2 A0A383YRE3 A0A1W5A6I2 G5ARS1 A0A2Y9DEG4
PDB
6GAZ
E-value=1.02532e-28,
Score=309
Ontologies
GO
Topology
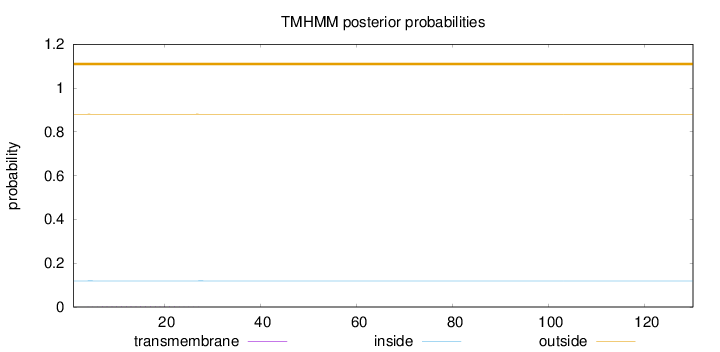
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02723
Exp number, first 60 AAs:
0.02703
Total prob of N-in:
0.11914
outside
1 - 130
Population Genetic Test Statistics
Pi
227.726991
Theta
166.95103
Tajima's D
1.144967
CLR
0.282783
CSRT
0.701114944252787
Interpretation
Uncertain
