Gene
KWMTBOMO09230 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003412
Annotation
ribosomal_protein_P1_[Bombyx_mori]
Full name
60S acidic ribosomal protein P1
Alternative Name
Acidic ribosomal protein RPA2
RP21C
RP21C
Location in the cell
Cytoplasmic Reliability : 1.925
Sequence
CDS
ATGGTGTCAAAAGCTGAATTAGCATGTGTTTACTCTGCTCTCATCCTGGTTGATGATGATGTTGCCGTAACTGGTGAGAAAATTTCCACCATCTTGAAAGCGGCGGCTGTAGATGTAGAGCCATATTGGCCAGGTCTGTTCGCCAAAGCCTTGGAAGGCATCAATGTCCGTGACCTGATCACCAACATCGGCTCTGGAGTGGGTGCTGCTCCGGCCGCTGGTGGAGTGCCAGCCGCTGCTGCTGCAGCCGGCGCTCCGGCCGAGGAGAAGAAGGAAGAAAAGAAAGAGGAGGAACCCGAGGAGTCCGACGATGACATGGGCTTCGGTCTCTTTGACTAA
Protein
MVSKAELACVYSALILVDDDVAVTGEKISTILKAAAVDVEPYWPGLFAKALEGINVRDLITNIGSGVGAAPAAGGVPAAAAAAGAPAEEKKEEKKEEEPEESDDDMGFGLFD
Summary
Subunit
P1 and P2 exist as dimers at the large ribosomal subunit.
Similarity
Belongs to the eukaryotic ribosomal protein P1/P2 family.
Keywords
Complete proteome
Phosphoprotein
Reference proteome
Ribonucleoprotein
Ribosomal protein
Feature
chain 60S acidic ribosomal protein P1
Uniprot
Q5UAU0
G0ZEC3
H9J1M5
I4DLW8
I4DIK7
A0A1E1WDA7
+ More
D1LYR7 A0A212EXT2 A0A3S2NGL3 E3UUV9 Q8WQJ1 A0A0L7KVA4 Q5MGJ5 A0A0A1XDQ4 E7DZ68 A0A194RK20 A0A2A4KAR6 Q6F454 B4LTL2 A0A0M3QTA0 B4KFI9 A2IAC4 W0C095 W0C3P6 W0C2P7 A0A1W4UQ67 M9PBK5 B4ICW8 B3N7Q2 B4Q638 P08570 K7IWC5 W8C3I2 A8CW94 A0A0K8U714 T1GDU0 Q29N41 B4G8A2 B0WU22 A0A1L8EIE2 O96938 A0A1Q3FB58 A0A1L8DS83 B4N0U4 A0A0P4VKR9 R4G8I3 A6YPP2 G3CJS4 A0A224Y1J2 A0A0V0G4T8 A0A069DVP3 U5EZ48 A0A158NU13 A0A194PLG0 R4UV70 A0A232FK66 A0A2P8XSK5 D1FPL7 A0A034VUK6 B4JDP3 Q1HRP7 A0A023EES4 Q6XIN9 V9IIV6 A0A088AHE1 A0A1B6C9L9 A0A411G5Y0 C4N193 B3MUH8 A0A1B6JCU6 A0A1B6G0G4 Q17BC2 A0A182QAP0 A0A0X7YHM7 A0A336KAW1 A0A1B6JAP6 A0A146L4M7 A0A1B6KCD2 A0A026WMK5 A0A182NTH5 A0A182JQL6 A0A182PCX7 A0A182U6L8 A0A182V6B4 A0A182L2Q8 A0A182X1D6 Q7PNA9 A0A182IEB6
D1LYR7 A0A212EXT2 A0A3S2NGL3 E3UUV9 Q8WQJ1 A0A0L7KVA4 Q5MGJ5 A0A0A1XDQ4 E7DZ68 A0A194RK20 A0A2A4KAR6 Q6F454 B4LTL2 A0A0M3QTA0 B4KFI9 A2IAC4 W0C095 W0C3P6 W0C2P7 A0A1W4UQ67 M9PBK5 B4ICW8 B3N7Q2 B4Q638 P08570 K7IWC5 W8C3I2 A8CW94 A0A0K8U714 T1GDU0 Q29N41 B4G8A2 B0WU22 A0A1L8EIE2 O96938 A0A1Q3FB58 A0A1L8DS83 B4N0U4 A0A0P4VKR9 R4G8I3 A6YPP2 G3CJS4 A0A224Y1J2 A0A0V0G4T8 A0A069DVP3 U5EZ48 A0A158NU13 A0A194PLG0 R4UV70 A0A232FK66 A0A2P8XSK5 D1FPL7 A0A034VUK6 B4JDP3 Q1HRP7 A0A023EES4 Q6XIN9 V9IIV6 A0A088AHE1 A0A1B6C9L9 A0A411G5Y0 C4N193 B3MUH8 A0A1B6JCU6 A0A1B6G0G4 Q17BC2 A0A182QAP0 A0A0X7YHM7 A0A336KAW1 A0A1B6JAP6 A0A146L4M7 A0A1B6KCD2 A0A026WMK5 A0A182NTH5 A0A182JQL6 A0A182PCX7 A0A182U6L8 A0A182V6B4 A0A182L2Q8 A0A182X1D6 Q7PNA9 A0A182IEB6
Pubmed
19121390
22651552
22118469
14668217
26263512
26227816
+ More
16023793 25830018 26354079 17994087 17437641 24152112 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 3122177 8501137 12537569 18327897 20075255 24495485 18194529 15632085 9931511 27129103 18207082 21058630 26334808 21347285 28648823 29403074 20441151 25348373 17204158 17510324 24945155 26483478 14525923 17550304 19576987 26823975 24508170 20966253 12364791 14747013 17210077
16023793 25830018 26354079 17994087 17437641 24152112 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 3122177 8501137 12537569 18327897 20075255 24495485 18194529 15632085 9931511 27129103 18207082 21058630 26334808 21347285 28648823 29403074 20441151 25348373 17204158 17510324 24945155 26483478 14525923 17550304 19576987 26823975 24508170 20966253 12364791 14747013 17210077
EMBL
AY769268
AAV34810.1
JF265001
AEL28832.1
BABH01023826
AK402286
+ More
BAM18908.1 AK401125 BAM17747.1 GDQN01006086 JAT84968.1 GU084341 ACY95374.1 AGBW02011690 OWR46295.1 RSAL01000112 RVE47037.1 HM590399 ADP21468.1 AY072285 KT218665 ODYU01005012 AAL62466.1 ALA09385.1 SOQ45481.1 JTDY01005421 KOB67016.1 AY829791 AAV91405.1 GBXI01004763 JAD09529.1 HQ424771 ADT80715.1 KQ460106 KPJ17779.1 NWSH01000012 PCG80883.1 AB180436 BAD26680.1 CH940649 EDW64985.1 CP012523 ALC38519.1 CH933807 EDW13104.1 EF179438 ABM55444.1 KC623037 KC623038 KC623039 KC623040 KC623041 KC623042 KC623043 KC623044 KC623045 KC623046 KC623047 KC623048 KC623049 KC623050 KC623051 KC623052 AHE75829.1 AHE75830.1 AHE75831.1 AHE75832.1 AHE75833.1 AHE75834.1 AHE75835.1 AHE75836.1 AHE75837.1 AHE75838.1 AHE75839.1 AHE75840.1 AHE75841.1 AHE75842.1 AHE75843.1 AHE75844.1 KC623053 KC623054 KC623055 KC623056 KC623057 KC623058 KC623059 KC623060 KC623061 KC623062 KC623063 KC623064 KC623065 KC623066 KC623067 AHE75845.1 AHE75846.1 AHE75847.1 AHE75848.1 AHE75849.1 AHE75850.1 AHE75851.1 AHE75852.1 AHE75853.1 AHE75854.1 AHE75855.1 AHE75856.1 AHE75857.1 AHE75858.1 AHE75859.1 KC623068 OUUW01000004 AHE75860.1 SPP79702.1 AE014134 AGB92359.1 CH480829 EDW45394.1 CH954177 EDV57228.1 CM000361 CM002910 EDX03222.1 KMY87310.1 Y00504 S62170 AY069125 GAMC01000068 JAC06488.1 EU124600 ABV60318.1 GDHF01030169 JAI22145.1 CAQQ02029573 CH379060 EAL33502.1 CH479180 EDW28582.1 DS232099 EDS34738.1 GFDG01000361 JAV18438.1 Y11907 CAA72658.1 GFDL01010293 JAV24752.1 GFDF01004785 JAV09299.1 CH963920 EDW77707.1 GDKW01000904 JAI55691.1 ACPB03013963 GAHY01000922 JAA76588.1 EF639053 GEMB01000142 ABR27938.1 JAS02971.1 HP639841 AEM97978.1 GFTR01001933 JAW14493.1 GECL01003730 JAP02394.1 GBGD01003535 JAC85354.1 GANO01001439 JAB58432.1 ADTU01002832 KQ459602 KPI93569.1 KC632327 AGM32141.1 NNAY01000110 OXU30889.1 PYGN01001417 PSN34993.1 EZ419767 ACY69940.1 GAKP01013417 GAKP01013413 JAC45535.1 CH916368 EDW03413.1 DQ440047 CH477274 ABF18080.1 EAT45193.1 JXUM01001213 JXUM01030309 GAPW01006252 KQ560880 JAC07346.1 KXJ80503.1 AY231791 CM000157 AAR09814.1 EDW87138.1 JR048019 AEY60557.1 GEDC01027269 JAS10029.1 MH365495 QBB01304.1 EZ048866 ACN69158.1 CH902624 EDV33507.1 GECU01010694 JAS97012.1 GECZ01013853 JAS55916.1 CH477324 EAT43524.1 AXCN02000703 KC507366 AHB12489.1 UFQS01000177 UFQT01000177 UFQT01000278 SSX00798.1 SSX21178.1 SSX22681.1 GECU01011521 JAS96185.1 GDHC01015236 JAQ03393.1 GEBQ01030870 GEBQ01006011 JAT09107.1 JAT33966.1 KK107167 EZA56349.1 AAAB01008964 EAA12468.5 APCN01005200
BAM18908.1 AK401125 BAM17747.1 GDQN01006086 JAT84968.1 GU084341 ACY95374.1 AGBW02011690 OWR46295.1 RSAL01000112 RVE47037.1 HM590399 ADP21468.1 AY072285 KT218665 ODYU01005012 AAL62466.1 ALA09385.1 SOQ45481.1 JTDY01005421 KOB67016.1 AY829791 AAV91405.1 GBXI01004763 JAD09529.1 HQ424771 ADT80715.1 KQ460106 KPJ17779.1 NWSH01000012 PCG80883.1 AB180436 BAD26680.1 CH940649 EDW64985.1 CP012523 ALC38519.1 CH933807 EDW13104.1 EF179438 ABM55444.1 KC623037 KC623038 KC623039 KC623040 KC623041 KC623042 KC623043 KC623044 KC623045 KC623046 KC623047 KC623048 KC623049 KC623050 KC623051 KC623052 AHE75829.1 AHE75830.1 AHE75831.1 AHE75832.1 AHE75833.1 AHE75834.1 AHE75835.1 AHE75836.1 AHE75837.1 AHE75838.1 AHE75839.1 AHE75840.1 AHE75841.1 AHE75842.1 AHE75843.1 AHE75844.1 KC623053 KC623054 KC623055 KC623056 KC623057 KC623058 KC623059 KC623060 KC623061 KC623062 KC623063 KC623064 KC623065 KC623066 KC623067 AHE75845.1 AHE75846.1 AHE75847.1 AHE75848.1 AHE75849.1 AHE75850.1 AHE75851.1 AHE75852.1 AHE75853.1 AHE75854.1 AHE75855.1 AHE75856.1 AHE75857.1 AHE75858.1 AHE75859.1 KC623068 OUUW01000004 AHE75860.1 SPP79702.1 AE014134 AGB92359.1 CH480829 EDW45394.1 CH954177 EDV57228.1 CM000361 CM002910 EDX03222.1 KMY87310.1 Y00504 S62170 AY069125 GAMC01000068 JAC06488.1 EU124600 ABV60318.1 GDHF01030169 JAI22145.1 CAQQ02029573 CH379060 EAL33502.1 CH479180 EDW28582.1 DS232099 EDS34738.1 GFDG01000361 JAV18438.1 Y11907 CAA72658.1 GFDL01010293 JAV24752.1 GFDF01004785 JAV09299.1 CH963920 EDW77707.1 GDKW01000904 JAI55691.1 ACPB03013963 GAHY01000922 JAA76588.1 EF639053 GEMB01000142 ABR27938.1 JAS02971.1 HP639841 AEM97978.1 GFTR01001933 JAW14493.1 GECL01003730 JAP02394.1 GBGD01003535 JAC85354.1 GANO01001439 JAB58432.1 ADTU01002832 KQ459602 KPI93569.1 KC632327 AGM32141.1 NNAY01000110 OXU30889.1 PYGN01001417 PSN34993.1 EZ419767 ACY69940.1 GAKP01013417 GAKP01013413 JAC45535.1 CH916368 EDW03413.1 DQ440047 CH477274 ABF18080.1 EAT45193.1 JXUM01001213 JXUM01030309 GAPW01006252 KQ560880 JAC07346.1 KXJ80503.1 AY231791 CM000157 AAR09814.1 EDW87138.1 JR048019 AEY60557.1 GEDC01027269 JAS10029.1 MH365495 QBB01304.1 EZ048866 ACN69158.1 CH902624 EDV33507.1 GECU01010694 JAS97012.1 GECZ01013853 JAS55916.1 CH477324 EAT43524.1 AXCN02000703 KC507366 AHB12489.1 UFQS01000177 UFQT01000177 UFQT01000278 SSX00798.1 SSX21178.1 SSX22681.1 GECU01011521 JAS96185.1 GDHC01015236 JAQ03393.1 GEBQ01030870 GEBQ01006011 JAT09107.1 JAT33966.1 KK107167 EZA56349.1 AAAB01008964 EAA12468.5 APCN01005200
Proteomes
UP000005204
UP000007151
UP000283053
UP000037510
UP000053240
UP000218220
+ More
UP000008792 UP000092553 UP000009192 UP000268350 UP000192221 UP000000803 UP000001292 UP000008711 UP000000304 UP000002358 UP000015102 UP000001819 UP000008744 UP000002320 UP000007798 UP000015103 UP000005205 UP000053268 UP000215335 UP000245037 UP000001070 UP000008820 UP000069940 UP000249989 UP000002282 UP000005203 UP000095300 UP000007801 UP000075886 UP000053097 UP000075884 UP000075881 UP000075885 UP000075902 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840
UP000008792 UP000092553 UP000009192 UP000268350 UP000192221 UP000000803 UP000001292 UP000008711 UP000000304 UP000002358 UP000015102 UP000001819 UP000008744 UP000002320 UP000007798 UP000015103 UP000005205 UP000053268 UP000215335 UP000245037 UP000001070 UP000008820 UP000069940 UP000249989 UP000002282 UP000005203 UP000095300 UP000007801 UP000075886 UP000053097 UP000075884 UP000075881 UP000075885 UP000075902 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840
Pfam
PF14631 FancD2
Interpro
Gene 3D
ProteinModelPortal
Q5UAU0
G0ZEC3
H9J1M5
I4DLW8
I4DIK7
A0A1E1WDA7
+ More
D1LYR7 A0A212EXT2 A0A3S2NGL3 E3UUV9 Q8WQJ1 A0A0L7KVA4 Q5MGJ5 A0A0A1XDQ4 E7DZ68 A0A194RK20 A0A2A4KAR6 Q6F454 B4LTL2 A0A0M3QTA0 B4KFI9 A2IAC4 W0C095 W0C3P6 W0C2P7 A0A1W4UQ67 M9PBK5 B4ICW8 B3N7Q2 B4Q638 P08570 K7IWC5 W8C3I2 A8CW94 A0A0K8U714 T1GDU0 Q29N41 B4G8A2 B0WU22 A0A1L8EIE2 O96938 A0A1Q3FB58 A0A1L8DS83 B4N0U4 A0A0P4VKR9 R4G8I3 A6YPP2 G3CJS4 A0A224Y1J2 A0A0V0G4T8 A0A069DVP3 U5EZ48 A0A158NU13 A0A194PLG0 R4UV70 A0A232FK66 A0A2P8XSK5 D1FPL7 A0A034VUK6 B4JDP3 Q1HRP7 A0A023EES4 Q6XIN9 V9IIV6 A0A088AHE1 A0A1B6C9L9 A0A411G5Y0 C4N193 B3MUH8 A0A1B6JCU6 A0A1B6G0G4 Q17BC2 A0A182QAP0 A0A0X7YHM7 A0A336KAW1 A0A1B6JAP6 A0A146L4M7 A0A1B6KCD2 A0A026WMK5 A0A182NTH5 A0A182JQL6 A0A182PCX7 A0A182U6L8 A0A182V6B4 A0A182L2Q8 A0A182X1D6 Q7PNA9 A0A182IEB6
D1LYR7 A0A212EXT2 A0A3S2NGL3 E3UUV9 Q8WQJ1 A0A0L7KVA4 Q5MGJ5 A0A0A1XDQ4 E7DZ68 A0A194RK20 A0A2A4KAR6 Q6F454 B4LTL2 A0A0M3QTA0 B4KFI9 A2IAC4 W0C095 W0C3P6 W0C2P7 A0A1W4UQ67 M9PBK5 B4ICW8 B3N7Q2 B4Q638 P08570 K7IWC5 W8C3I2 A8CW94 A0A0K8U714 T1GDU0 Q29N41 B4G8A2 B0WU22 A0A1L8EIE2 O96938 A0A1Q3FB58 A0A1L8DS83 B4N0U4 A0A0P4VKR9 R4G8I3 A6YPP2 G3CJS4 A0A224Y1J2 A0A0V0G4T8 A0A069DVP3 U5EZ48 A0A158NU13 A0A194PLG0 R4UV70 A0A232FK66 A0A2P8XSK5 D1FPL7 A0A034VUK6 B4JDP3 Q1HRP7 A0A023EES4 Q6XIN9 V9IIV6 A0A088AHE1 A0A1B6C9L9 A0A411G5Y0 C4N193 B3MUH8 A0A1B6JCU6 A0A1B6G0G4 Q17BC2 A0A182QAP0 A0A0X7YHM7 A0A336KAW1 A0A1B6JAP6 A0A146L4M7 A0A1B6KCD2 A0A026WMK5 A0A182NTH5 A0A182JQL6 A0A182PCX7 A0A182U6L8 A0A182V6B4 A0A182L2Q8 A0A182X1D6 Q7PNA9 A0A182IEB6
PDB
2LBF
E-value=3.91304e-17,
Score=209
Ontologies
GO
PANTHER
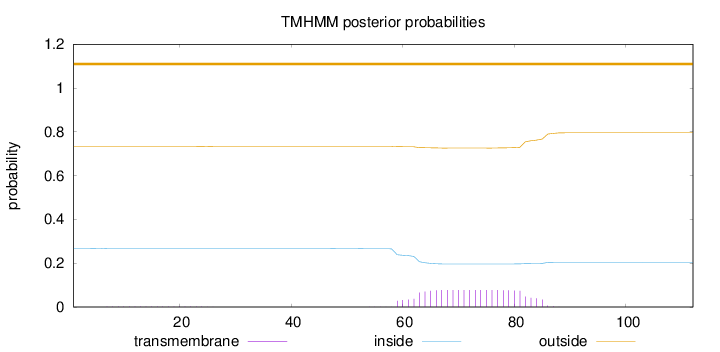
Topology
Length:
112
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.77293
Exp number, first 60 AAs:
0.0919
Total prob of N-in:
0.26783
outside
1 - 112
Population Genetic Test Statistics
Pi
237.035614
Theta
193.570906
Tajima's D
0.706689
CLR
1.567085
CSRT
0.576671166441678
Interpretation
Uncertain
