Pre Gene Modal
BGIBMGA003340
Annotation
PREDICTED:_presqualene_diphosphate_phosphatase-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.844
Sequence
CDS
ATGGATATGTTAGGAGAAACAGAGAAAAAGCGTCAAACTCCACCCATGTTGAAAAAAATTTTGCAGTATGATGTTCAAATTACGAAGACATTTGTTGACAGAGCACTAAAAATAACTGCTCTAAAGTCTTTACGGAATCATTCTCAATTACTAGAGATATCATGTCATGGCATAGTTTGGCTTGCAGGCTGGCTCACTTTCATATGGCTGTTTAATAGTAAAGATCTTTACCAGTTGCAAGTTAATATGTTGATTGCACTAATTTTAGACATTGTTGTGGTAGCTGTATTGAAAGCATTTGTAAGAAGACGTCGACCGGTACCAATGAATAAGCTCATGGAAGTTGGTCCTGATAAGTTCTCATTCCCATCCGGACATGCAAGCAGGGCTGTGCTCATCTCGTTCATTTTAATATACTTTGATTCTGTATCAATAATTTTCTACCCCCCACTTATGGCTTGGGTAGTTTCAGTGTCCATTTCCAGGGTATTGGCTGAACGTCACTATCTACTAGACAGCATAGGAGGTGTGGGTATTGGAATTCTTGAAGGTCTCTTCATGTCCCTCATATGGTTGTCCCAAAGTACATCTGCTAGTATTCTGTCATCATTGTCTGATGAAAAATTAGATGGTGGAGAATACCATGTATAA
Protein
MDMLGETEKKRQTPPMLKKILQYDVQITKTFVDRALKITALKSLRNHSQLLEISCHGIVWLAGWLTFIWLFNSKDLYQLQVNMLIALILDIVVVAVLKAFVRRRRPVPMNKLMEVGPDKFSFPSGHASRAVLISFILIYFDSVSIIFYPPLMAWVVSVSISRVLAERHYLLDSIGGVGIGILEGLFMSLIWLSQSTSASILSSLSDEKLDGGEYHV
Summary
Uniprot
H9J1F3
A0A194PJS3
A0A437B9H6
A0A212EXU8
A0A0L7K3Y4
S4P1S1
+ More
A0A194RKB3 A0A2H1VX98 A0A1L8EE58 K7IME2 A0A1L8EE31 A0A084WDC5 A0A182XMF7 Q16Y29 A0A023EKA4 A0A182JA74 A0A023EMM7 A0A182UTN0 A0A182TK63 A0A182KQ20 Q7PNM7 A0A182QB05 A0A023EK54 A0A1I8NFV9 A0A182M2Z2 A0A182I287 A0A2M4CKB9 A0A2M3Z9F9 A0A2M4CKD6 W5JVB1 A0A2M4BYS7 A0A182NJG8 A0A2M3Z9N8 U5EUT2 A0A2M4BZQ7 A0A182RFH7 A0A2M4AJH8 A0A182FGV0 A0A2M4AJQ3 A0A182PIF4 A0A182YJA0 T1EAF6 A0A182WL87 A0A2J7QI07 D6WEE8 A0A1W4WJT3 A0A232FC98 A0A0C9R8H6 B0W3S1 A0A1Q3FG00 A0A1B6D5H9 A0A067RID5 B4HWI4 B4Q8R7 A0A1W4UST5 E2C5Y4 Q8T8T9 A0A336LDD4 B3MPN8 A0A023EI16 A0A1L8DYI9 B4NZ72 A0A1L8DYI4 B3N948 T1GI00 B4M8H1 A0A0T6AYM7 B4JPP4 A0A1I8PFI8 A0A3L8DE71 B4KHN9 A0A1B6I6W7 A0A0L0CFG9 A0A0Q9XI03 F4X0F0 A0A1A9WEV7 A0A0L7R9M5 E0VT09 A0A310S5I3 A0A158NFR3 A0A2A3E021 A0A088ADU5 V9IGP8 A0A1B6H0B0 A0A0C9RUE0 Q29NI1 A0A1J1IG68 A0A0K8VX00 B4G7H8 A0A026WRZ1 A0A1B6M417 A0A026W857 A0A151K1G1 A0A151WNZ5 J9K5Q9 T1D312 A0A2P8XG18 A0A195BDN9 A0A0K8W0Q0
A0A194RKB3 A0A2H1VX98 A0A1L8EE58 K7IME2 A0A1L8EE31 A0A084WDC5 A0A182XMF7 Q16Y29 A0A023EKA4 A0A182JA74 A0A023EMM7 A0A182UTN0 A0A182TK63 A0A182KQ20 Q7PNM7 A0A182QB05 A0A023EK54 A0A1I8NFV9 A0A182M2Z2 A0A182I287 A0A2M4CKB9 A0A2M3Z9F9 A0A2M4CKD6 W5JVB1 A0A2M4BYS7 A0A182NJG8 A0A2M3Z9N8 U5EUT2 A0A2M4BZQ7 A0A182RFH7 A0A2M4AJH8 A0A182FGV0 A0A2M4AJQ3 A0A182PIF4 A0A182YJA0 T1EAF6 A0A182WL87 A0A2J7QI07 D6WEE8 A0A1W4WJT3 A0A232FC98 A0A0C9R8H6 B0W3S1 A0A1Q3FG00 A0A1B6D5H9 A0A067RID5 B4HWI4 B4Q8R7 A0A1W4UST5 E2C5Y4 Q8T8T9 A0A336LDD4 B3MPN8 A0A023EI16 A0A1L8DYI9 B4NZ72 A0A1L8DYI4 B3N948 T1GI00 B4M8H1 A0A0T6AYM7 B4JPP4 A0A1I8PFI8 A0A3L8DE71 B4KHN9 A0A1B6I6W7 A0A0L0CFG9 A0A0Q9XI03 F4X0F0 A0A1A9WEV7 A0A0L7R9M5 E0VT09 A0A310S5I3 A0A158NFR3 A0A2A3E021 A0A088ADU5 V9IGP8 A0A1B6H0B0 A0A0C9RUE0 Q29NI1 A0A1J1IG68 A0A0K8VX00 B4G7H8 A0A026WRZ1 A0A1B6M417 A0A026W857 A0A151K1G1 A0A151WNZ5 J9K5Q9 T1D312 A0A2P8XG18 A0A195BDN9 A0A0K8W0Q0
Pubmed
19121390
26354079
22118469
26227816
23622113
20075255
+ More
24438588 17510324 24945155 20966253 12364791 14747013 17210077 25315136 20920257 23761445 25244985 18362917 19820115 28648823 24845553 17994087 22936249 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26108605 21719571 20566863 21347285 15632085 24508170 24330624 29403074
24438588 17510324 24945155 20966253 12364791 14747013 17210077 25315136 20920257 23761445 25244985 18362917 19820115 28648823 24845553 17994087 22936249 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 26108605 21719571 20566863 21347285 15632085 24508170 24330624 29403074
EMBL
BABH01023818
BABH01023819
BABH01023820
KQ459602
KPI93567.1
RSAL01000112
+ More
RVE47041.1 AGBW02011690 OWR46291.1 JTDY01011271 KOB57161.1 GAIX01012345 JAA80215.1 KQ460106 KPJ17775.1 ODYU01005012 SOQ45477.1 GFDG01001818 JAV16981.1 GFDG01001819 JAV16980.1 ATLV01022986 KE525339 KFB48219.1 CH477526 EAT39531.1 GAPW01004030 JAC09568.1 GAPW01004009 JAC09589.1 AAAB01008960 EAA11926.4 AXCN02000446 GAPW01004090 JAC09508.1 AXCM01004988 APCN01000128 GGFL01001608 MBW65786.1 GGFM01004403 MBW25154.1 GGFL01001609 MBW65787.1 ADMH02000243 ETN67258.1 GGFJ01009078 MBW58219.1 GGFM01004480 MBW25231.1 GANO01003729 JAB56142.1 GGFJ01009077 MBW58218.1 GGFK01007593 MBW40914.1 GGFK01007680 MBW41001.1 GAMD01000462 JAB01129.1 NEVH01013964 PNF28183.1 KQ971318 EFA00398.1 NNAY01000431 OXU28444.1 GBYB01004380 JAG74147.1 DS231833 EDS32218.1 GFDL01008565 JAV26480.1 GEDC01016473 JAS20825.1 KK852458 KDR23552.1 CH480818 EDW52379.1 CM000361 CM002910 EDX04482.1 KMY89448.1 GL452875 EFN76643.1 AY075278 AE014134 AAL68145.1 AAN10729.1 AGB92852.1 UFQS01003315 UFQT01002298 UFQT01003315 SSX15479.1 SSX33146.1 CH902620 EDV32286.1 GAPW01004575 JAC09023.1 GFDF01002597 JAV11487.1 CM000157 EDW88767.1 GFDF01002598 JAV11486.1 CH954177 EDV58483.2 CAQQ02390494 CH940654 EDW57497.2 LJIG01022599 KRT79719.1 CH916372 EDV98874.1 QOIP01000009 RLU18780.1 CH933807 EDW12318.2 GECU01030620 GECU01025035 JAS77086.1 JAS82671.1 JRES01000473 KNC30986.1 KRG03187.1 GL888497 EGI60108.1 KQ414627 KOC67456.1 DS235759 EEB16515.1 KQ769940 OAD52747.1 ADTU01014582 KZ288515 PBC25110.1 JR044183 AEY59847.1 GECZ01001645 JAS68124.1 GBYB01012440 JAG82207.1 CH379059 EAL34239.3 CVRI01000048 CRK98762.1 GDHF01012441 GDHF01008912 JAI39873.1 JAI43402.1 CH479180 EDW28378.1 KK107119 EZA58792.1 GEBQ01009296 JAT30681.1 KK107348 EZA52143.1 KQ981163 KYN45395.1 KQ982893 KYQ49550.1 ABLF02034274 GALA01001428 JAA93424.1 PYGN01002270 PSN30949.1 KQ976514 KYM82322.1 GDHF01007894 JAI44420.1
RVE47041.1 AGBW02011690 OWR46291.1 JTDY01011271 KOB57161.1 GAIX01012345 JAA80215.1 KQ460106 KPJ17775.1 ODYU01005012 SOQ45477.1 GFDG01001818 JAV16981.1 GFDG01001819 JAV16980.1 ATLV01022986 KE525339 KFB48219.1 CH477526 EAT39531.1 GAPW01004030 JAC09568.1 GAPW01004009 JAC09589.1 AAAB01008960 EAA11926.4 AXCN02000446 GAPW01004090 JAC09508.1 AXCM01004988 APCN01000128 GGFL01001608 MBW65786.1 GGFM01004403 MBW25154.1 GGFL01001609 MBW65787.1 ADMH02000243 ETN67258.1 GGFJ01009078 MBW58219.1 GGFM01004480 MBW25231.1 GANO01003729 JAB56142.1 GGFJ01009077 MBW58218.1 GGFK01007593 MBW40914.1 GGFK01007680 MBW41001.1 GAMD01000462 JAB01129.1 NEVH01013964 PNF28183.1 KQ971318 EFA00398.1 NNAY01000431 OXU28444.1 GBYB01004380 JAG74147.1 DS231833 EDS32218.1 GFDL01008565 JAV26480.1 GEDC01016473 JAS20825.1 KK852458 KDR23552.1 CH480818 EDW52379.1 CM000361 CM002910 EDX04482.1 KMY89448.1 GL452875 EFN76643.1 AY075278 AE014134 AAL68145.1 AAN10729.1 AGB92852.1 UFQS01003315 UFQT01002298 UFQT01003315 SSX15479.1 SSX33146.1 CH902620 EDV32286.1 GAPW01004575 JAC09023.1 GFDF01002597 JAV11487.1 CM000157 EDW88767.1 GFDF01002598 JAV11486.1 CH954177 EDV58483.2 CAQQ02390494 CH940654 EDW57497.2 LJIG01022599 KRT79719.1 CH916372 EDV98874.1 QOIP01000009 RLU18780.1 CH933807 EDW12318.2 GECU01030620 GECU01025035 JAS77086.1 JAS82671.1 JRES01000473 KNC30986.1 KRG03187.1 GL888497 EGI60108.1 KQ414627 KOC67456.1 DS235759 EEB16515.1 KQ769940 OAD52747.1 ADTU01014582 KZ288515 PBC25110.1 JR044183 AEY59847.1 GECZ01001645 JAS68124.1 GBYB01012440 JAG82207.1 CH379059 EAL34239.3 CVRI01000048 CRK98762.1 GDHF01012441 GDHF01008912 JAI39873.1 JAI43402.1 CH479180 EDW28378.1 KK107119 EZA58792.1 GEBQ01009296 JAT30681.1 KK107348 EZA52143.1 KQ981163 KYN45395.1 KQ982893 KYQ49550.1 ABLF02034274 GALA01001428 JAA93424.1 PYGN01002270 PSN30949.1 KQ976514 KYM82322.1 GDHF01007894 JAI44420.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000037510
UP000053240
+ More
UP000002358 UP000030765 UP000076407 UP000008820 UP000075880 UP000075903 UP000075902 UP000075882 UP000007062 UP000075886 UP000095301 UP000075883 UP000075840 UP000000673 UP000075884 UP000075900 UP000069272 UP000075885 UP000076408 UP000075920 UP000235965 UP000007266 UP000192223 UP000215335 UP000002320 UP000027135 UP000001292 UP000000304 UP000192221 UP000008237 UP000000803 UP000007801 UP000002282 UP000008711 UP000015102 UP000008792 UP000001070 UP000095300 UP000279307 UP000009192 UP000037069 UP000007755 UP000091820 UP000053825 UP000009046 UP000005205 UP000242457 UP000005203 UP000001819 UP000183832 UP000008744 UP000053097 UP000078541 UP000075809 UP000007819 UP000245037 UP000078540
UP000002358 UP000030765 UP000076407 UP000008820 UP000075880 UP000075903 UP000075902 UP000075882 UP000007062 UP000075886 UP000095301 UP000075883 UP000075840 UP000000673 UP000075884 UP000075900 UP000069272 UP000075885 UP000076408 UP000075920 UP000235965 UP000007266 UP000192223 UP000215335 UP000002320 UP000027135 UP000001292 UP000000304 UP000192221 UP000008237 UP000000803 UP000007801 UP000002282 UP000008711 UP000015102 UP000008792 UP000001070 UP000095300 UP000279307 UP000009192 UP000037069 UP000007755 UP000091820 UP000053825 UP000009046 UP000005205 UP000242457 UP000005203 UP000001819 UP000183832 UP000008744 UP000053097 UP000078541 UP000075809 UP000007819 UP000245037 UP000078540
Interpro
ProteinModelPortal
H9J1F3
A0A194PJS3
A0A437B9H6
A0A212EXU8
A0A0L7K3Y4
S4P1S1
+ More
A0A194RKB3 A0A2H1VX98 A0A1L8EE58 K7IME2 A0A1L8EE31 A0A084WDC5 A0A182XMF7 Q16Y29 A0A023EKA4 A0A182JA74 A0A023EMM7 A0A182UTN0 A0A182TK63 A0A182KQ20 Q7PNM7 A0A182QB05 A0A023EK54 A0A1I8NFV9 A0A182M2Z2 A0A182I287 A0A2M4CKB9 A0A2M3Z9F9 A0A2M4CKD6 W5JVB1 A0A2M4BYS7 A0A182NJG8 A0A2M3Z9N8 U5EUT2 A0A2M4BZQ7 A0A182RFH7 A0A2M4AJH8 A0A182FGV0 A0A2M4AJQ3 A0A182PIF4 A0A182YJA0 T1EAF6 A0A182WL87 A0A2J7QI07 D6WEE8 A0A1W4WJT3 A0A232FC98 A0A0C9R8H6 B0W3S1 A0A1Q3FG00 A0A1B6D5H9 A0A067RID5 B4HWI4 B4Q8R7 A0A1W4UST5 E2C5Y4 Q8T8T9 A0A336LDD4 B3MPN8 A0A023EI16 A0A1L8DYI9 B4NZ72 A0A1L8DYI4 B3N948 T1GI00 B4M8H1 A0A0T6AYM7 B4JPP4 A0A1I8PFI8 A0A3L8DE71 B4KHN9 A0A1B6I6W7 A0A0L0CFG9 A0A0Q9XI03 F4X0F0 A0A1A9WEV7 A0A0L7R9M5 E0VT09 A0A310S5I3 A0A158NFR3 A0A2A3E021 A0A088ADU5 V9IGP8 A0A1B6H0B0 A0A0C9RUE0 Q29NI1 A0A1J1IG68 A0A0K8VX00 B4G7H8 A0A026WRZ1 A0A1B6M417 A0A026W857 A0A151K1G1 A0A151WNZ5 J9K5Q9 T1D312 A0A2P8XG18 A0A195BDN9 A0A0K8W0Q0
A0A194RKB3 A0A2H1VX98 A0A1L8EE58 K7IME2 A0A1L8EE31 A0A084WDC5 A0A182XMF7 Q16Y29 A0A023EKA4 A0A182JA74 A0A023EMM7 A0A182UTN0 A0A182TK63 A0A182KQ20 Q7PNM7 A0A182QB05 A0A023EK54 A0A1I8NFV9 A0A182M2Z2 A0A182I287 A0A2M4CKB9 A0A2M3Z9F9 A0A2M4CKD6 W5JVB1 A0A2M4BYS7 A0A182NJG8 A0A2M3Z9N8 U5EUT2 A0A2M4BZQ7 A0A182RFH7 A0A2M4AJH8 A0A182FGV0 A0A2M4AJQ3 A0A182PIF4 A0A182YJA0 T1EAF6 A0A182WL87 A0A2J7QI07 D6WEE8 A0A1W4WJT3 A0A232FC98 A0A0C9R8H6 B0W3S1 A0A1Q3FG00 A0A1B6D5H9 A0A067RID5 B4HWI4 B4Q8R7 A0A1W4UST5 E2C5Y4 Q8T8T9 A0A336LDD4 B3MPN8 A0A023EI16 A0A1L8DYI9 B4NZ72 A0A1L8DYI4 B3N948 T1GI00 B4M8H1 A0A0T6AYM7 B4JPP4 A0A1I8PFI8 A0A3L8DE71 B4KHN9 A0A1B6I6W7 A0A0L0CFG9 A0A0Q9XI03 F4X0F0 A0A1A9WEV7 A0A0L7R9M5 E0VT09 A0A310S5I3 A0A158NFR3 A0A2A3E021 A0A088ADU5 V9IGP8 A0A1B6H0B0 A0A0C9RUE0 Q29NI1 A0A1J1IG68 A0A0K8VX00 B4G7H8 A0A026WRZ1 A0A1B6M417 A0A026W857 A0A151K1G1 A0A151WNZ5 J9K5Q9 T1D312 A0A2P8XG18 A0A195BDN9 A0A0K8W0Q0
Ontologies
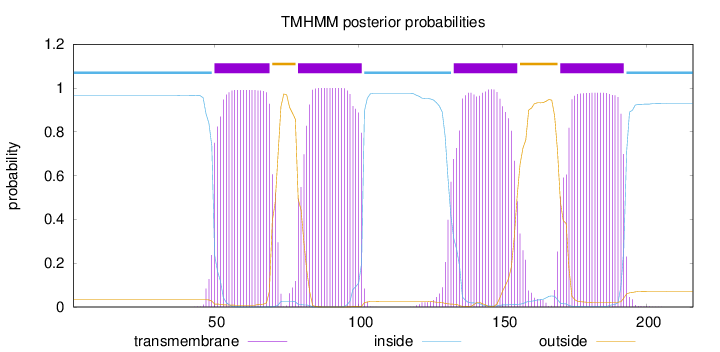
Topology
Length:
216
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.79536
Exp number, first 60 AAs:
10.75679
Total prob of N-in:
0.96672
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 69
outside
70 - 78
TMhelix
79 - 101
inside
102 - 132
TMhelix
133 - 155
outside
156 - 169
TMhelix
170 - 192
inside
193 - 216
Population Genetic Test Statistics
Pi
492.611576
Theta
202.903007
Tajima's D
4.543254
CLR
0.000881
CSRT
0.999850007499625
Interpretation
Uncertain
