Gene
KWMTBOMO09227
Annotation
PREDICTED:_thiamine_transporter_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.912
Sequence
CDS
ATGTACATGGCCACTGAAGTTGCTTATTACACTTACATTTATGCTAAGGTGGATCCGAAGAAGTACCCTTTAGTTACGTCGTGTACTAGGATCGCATCATTGACAGGAAGGTTCCTATCTGGTGTCAGTTCACAACTACTCATACATTTCAAATTCATGGACTACAGGGAACTAAATTACATAACTTTGACAGCGCAGATACTAGCAACATTTTGGGCACTATGGCTGCCTCCGGTTAAATATGGAATATACTTTCACAGAAGGATATCGACAATTAATATCCCTGTTGACAAATCATCATATAAAAACTCAGAAAAGCGTACATTCACAAGTAATTTAATAGAGGCTTCGAGCTTAATATTGAAACACTCGAGGGCGGCTTACTCCCGTCCAAAAGTGATAGCGTGGAGCGCTCTGTATGCGGTCGCTTTAGCCCTCTTCGTGCAAGCTCAGACTTATATACAAATACTGTGGCGGTACATACAAAAAGAACAATCGAATCCGGTATTATACAATGGAGCTGCGGAAGCCGCTACCACTCTGTTAGGGGCCGGGGCTGCGTACATAGCATCGTATGACCGGCAGTCCTTCAGTCCCGCACTTTCTTCAGCCATACAAGGAATCGCTCTGTTTCTAACTACCTACATATCGAACGTATTCGTTTCCTACGCTGGATACGTTGTCATGGGAGTCATGTTTCATTACATCATCACTTTGGCCAGCGCAAAAATAGCTAGTCAATTAATCGACGACAGTTGTTTCGGGCTGATATTCGGTATCAACACACTCCTCGGCGTCGCTTTGCAGTCTCTCCTCACGTACATACTGTTACAAAACTTGGAGTTTCACATAACAACGCACTATTACATCGTAAGCGGTCTTTTTATAACTCTGGCCACTTGTTGGTTAGTCGGATGGATACTCCATATTGCTCATAATAAAAAACAAAACGTGAACAGTAATTAA
Protein
MYMATEVAYYTYIYAKVDPKKYPLVTSCTRIASLTGRFLSGVSSQLLIHFKFMDYRELNYITLTAQILATFWALWLPPVKYGIYFHRRISTINIPVDKSSYKNSEKRTFTSNLIEASSLILKHSRAAYSRPKVIAWSALYAVALALFVQAQTYIQILWRYIQKEQSNPVLYNGAAEAATTLLGAGAAYIASYDRQSFSPALSSAIQGIALFLTTYISNVFVSYAGYVVMGVMFHYIITLASAKIASQLIDDSCFGLIFGINTLLGVALQSLLTYILLQNLEFHITTHYYIVSGLFITLATCWLVGWILHIAHNKKQNVNSN
Summary
Similarity
Belongs to the serpin family.
Uniprot
A0A2A4K9B9
A0A437B9I2
A0A194RIX0
A0A212EXR6
A0A1E1WJ69
A0A194PR11
+ More
A0A1A9X432 A0A1W4V4M3 A0A1B0FNL7 A0A1A9YFY6 A0A084VJC1 A0A1B0AF25 B4NKW1 A0A1A9VU41 A0A1B0BH53 A0A0L0C7D6 B3LZG0 A0A023ESB1 A0A3B0KBK3 A0A182G3B7 A0A1I8N4B8 A0A1L8E8F6 B4QUM9 A0A1L8DE13 A0A1L8E8M5 B4GLC6 A0A1L8E8P8 Q294A4 Q9VGV5 B3LZF8 B4HIN5 A0A1L8EFX5 A0A0M3QYC5 A0A1L8EFP6 A0A0K8U209 A0A0A1XPW3 B4JGE1 A0A1L8EFP2 A0A1I8PS62 B4K5T8 B4JGE0 B0WKU6 J9EAM5 B3P1I7 A0A1J1IHM5 A0A034VQF7 B4PLE7 A0A1W4V4Y9 B4LZX8 Q9VGV6 A0A182N673 B4HIN6 B4LZX7 A0A1Q3FH43 B4PLE6 D2A676 A0A182JKS1 A0A195DKU9 W8C661 A0A182QBU7 A0A182NKA2 B3LZF9 A0A151X988 B4QUN0 B0WCL0 B4K5T9 B3P1I8 A0A336M170 B4GLC5 A0A1B0G7A7 A0A182WCD3 Q294A5 A0A195CXW0 Q7Q5N2 B4NKW0 A0A026WFU5 A0A0M4EGB4 A0A182QPN5 A0A336MEJ3 A0A0K8UQZ4 A0A182G1M5 E2A357 D6WJH6 A0A151JVH3 B3MNJ2 A0A1A9UGV0 A0A1Y1KHU6 U4UKF2 E9IXH9 A0A1B0C4Z9 A0A336LUE0 N6UCU0 E9IYY4 U4U6M1 A0A3L8DKQ6 A0A1B0DN88 A0A1A9XUE4 A0A1J1IHU2
A0A1A9X432 A0A1W4V4M3 A0A1B0FNL7 A0A1A9YFY6 A0A084VJC1 A0A1B0AF25 B4NKW1 A0A1A9VU41 A0A1B0BH53 A0A0L0C7D6 B3LZG0 A0A023ESB1 A0A3B0KBK3 A0A182G3B7 A0A1I8N4B8 A0A1L8E8F6 B4QUM9 A0A1L8DE13 A0A1L8E8M5 B4GLC6 A0A1L8E8P8 Q294A4 Q9VGV5 B3LZF8 B4HIN5 A0A1L8EFX5 A0A0M3QYC5 A0A1L8EFP6 A0A0K8U209 A0A0A1XPW3 B4JGE1 A0A1L8EFP2 A0A1I8PS62 B4K5T8 B4JGE0 B0WKU6 J9EAM5 B3P1I7 A0A1J1IHM5 A0A034VQF7 B4PLE7 A0A1W4V4Y9 B4LZX8 Q9VGV6 A0A182N673 B4HIN6 B4LZX7 A0A1Q3FH43 B4PLE6 D2A676 A0A182JKS1 A0A195DKU9 W8C661 A0A182QBU7 A0A182NKA2 B3LZF9 A0A151X988 B4QUN0 B0WCL0 B4K5T9 B3P1I8 A0A336M170 B4GLC5 A0A1B0G7A7 A0A182WCD3 Q294A5 A0A195CXW0 Q7Q5N2 B4NKW0 A0A026WFU5 A0A0M4EGB4 A0A182QPN5 A0A336MEJ3 A0A0K8UQZ4 A0A182G1M5 E2A357 D6WJH6 A0A151JVH3 B3MNJ2 A0A1A9UGV0 A0A1Y1KHU6 U4UKF2 E9IXH9 A0A1B0C4Z9 A0A336LUE0 N6UCU0 E9IYY4 U4U6M1 A0A3L8DKQ6 A0A1B0DN88 A0A1A9XUE4 A0A1J1IHU2
Pubmed
26354079
22118469
24438588
17994087
26108605
24945155
+ More
26483478 25315136 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17510324 25348373 17550304 18362917 19820115 24495485 12364791 14747013 17210077 24508170 20798317 28004739 23537049 21282665 30249741
26483478 25315136 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17510324 25348373 17550304 18362917 19820115 24495485 12364791 14747013 17210077 24508170 20798317 28004739 23537049 21282665 30249741
EMBL
NWSH01000012
PCG80875.1
RSAL01000112
RVE47042.1
KQ460106
KPJ17773.1
+ More
AGBW02011690 OWR46289.1 GDQN01004076 JAT86978.1 KQ459602 KPI93565.1 CCAG010011488 ATLV01013503 KE524866 KFB38065.1 CH964272 EDW84164.1 JXJN01014183 JRES01000819 KNC28156.1 CH902617 EDV44139.1 GAPW01001465 JAC12133.1 OUUW01000007 SPP83509.1 JXUM01040706 KQ561256 KXJ79202.1 GFDG01003845 JAV14954.1 CM000364 EDX13436.1 GFDF01009504 JAV04580.1 GFDG01003846 JAV14953.1 CH479185 EDW38350.1 GFDG01003847 JAV14952.1 CM000070 EAL29060.1 KRT01265.1 AE014297 BT024295 AAF54568.1 ABC86357.1 EDV44137.2 CH480815 EDW42682.1 GFDG01001253 JAV17546.1 CP012526 ALC47379.1 GFDG01001252 JAV17547.1 GDHF01031736 JAI20578.1 GBXI01008316 GBXI01001397 JAD05976.1 JAD12895.1 CH916369 EDV92610.1 GFDG01001257 JAV17542.1 CH933806 EDW15150.1 EDV92609.1 DS231976 EDS30069.1 CH477621 EJY57841.1 CH954181 EDV49586.1 CVRI01000048 CRK99036.1 GAKP01014610 GAKP01014609 JAC44342.1 CM000160 EDW96854.1 CH940650 EDW67206.1 BT022488 AAF54567.2 AAY54904.1 EDW42683.1 EDW67205.1 GFDL01008191 JAV26854.1 EDW96853.1 KQ971346 EFA04966.1 KQ980765 KYN13471.1 GAMC01004459 JAC02097.1 AXCN02000270 EDV44138.1 KQ982383 KYQ56931.1 EDX13437.1 DS231889 EDS43661.1 EDW15151.1 EDV49587.1 UFQT01000411 SSX24066.1 EDW38349.1 CCAG010022298 EAL29059.2 KQ977141 KYN05417.1 AAAB01008960 EAA10851.4 EDW84163.2 KK107238 EZA54803.1 ALC47378.1 AXCN02000423 UFQT01000805 SSX27349.1 GDHF01023308 JAI29006.1 JXUM01137309 KQ568559 KXJ68959.1 GL436277 EFN72132.1 KQ971342 EFA03674.1 KQ981692 KYN37889.1 CH902620 EDV32100.1 GEZM01083921 JAV60944.1 KB632391 ERL94559.1 GL766755 EFZ14695.1 JXJN01014748 JXJN01025742 UFQT01000197 SSX21554.1 APGK01034125 KB740904 ENN78451.1 GL767077 EFZ14230.1 KB632006 ERL87963.1 QOIP01000007 RLU20489.1 AJVK01017384 CRK98614.1
AGBW02011690 OWR46289.1 GDQN01004076 JAT86978.1 KQ459602 KPI93565.1 CCAG010011488 ATLV01013503 KE524866 KFB38065.1 CH964272 EDW84164.1 JXJN01014183 JRES01000819 KNC28156.1 CH902617 EDV44139.1 GAPW01001465 JAC12133.1 OUUW01000007 SPP83509.1 JXUM01040706 KQ561256 KXJ79202.1 GFDG01003845 JAV14954.1 CM000364 EDX13436.1 GFDF01009504 JAV04580.1 GFDG01003846 JAV14953.1 CH479185 EDW38350.1 GFDG01003847 JAV14952.1 CM000070 EAL29060.1 KRT01265.1 AE014297 BT024295 AAF54568.1 ABC86357.1 EDV44137.2 CH480815 EDW42682.1 GFDG01001253 JAV17546.1 CP012526 ALC47379.1 GFDG01001252 JAV17547.1 GDHF01031736 JAI20578.1 GBXI01008316 GBXI01001397 JAD05976.1 JAD12895.1 CH916369 EDV92610.1 GFDG01001257 JAV17542.1 CH933806 EDW15150.1 EDV92609.1 DS231976 EDS30069.1 CH477621 EJY57841.1 CH954181 EDV49586.1 CVRI01000048 CRK99036.1 GAKP01014610 GAKP01014609 JAC44342.1 CM000160 EDW96854.1 CH940650 EDW67206.1 BT022488 AAF54567.2 AAY54904.1 EDW42683.1 EDW67205.1 GFDL01008191 JAV26854.1 EDW96853.1 KQ971346 EFA04966.1 KQ980765 KYN13471.1 GAMC01004459 JAC02097.1 AXCN02000270 EDV44138.1 KQ982383 KYQ56931.1 EDX13437.1 DS231889 EDS43661.1 EDW15151.1 EDV49587.1 UFQT01000411 SSX24066.1 EDW38349.1 CCAG010022298 EAL29059.2 KQ977141 KYN05417.1 AAAB01008960 EAA10851.4 EDW84163.2 KK107238 EZA54803.1 ALC47378.1 AXCN02000423 UFQT01000805 SSX27349.1 GDHF01023308 JAI29006.1 JXUM01137309 KQ568559 KXJ68959.1 GL436277 EFN72132.1 KQ971342 EFA03674.1 KQ981692 KYN37889.1 CH902620 EDV32100.1 GEZM01083921 JAV60944.1 KB632391 ERL94559.1 GL766755 EFZ14695.1 JXJN01014748 JXJN01025742 UFQT01000197 SSX21554.1 APGK01034125 KB740904 ENN78451.1 GL767077 EFZ14230.1 KB632006 ERL87963.1 QOIP01000007 RLU20489.1 AJVK01017384 CRK98614.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
UP000091820
+ More
UP000192221 UP000092444 UP000092443 UP000030765 UP000092445 UP000007798 UP000078200 UP000092460 UP000037069 UP000007801 UP000268350 UP000069940 UP000249989 UP000095301 UP000000304 UP000008744 UP000001819 UP000000803 UP000001292 UP000092553 UP000001070 UP000095300 UP000009192 UP000002320 UP000008820 UP000008711 UP000183832 UP000002282 UP000008792 UP000075884 UP000007266 UP000075880 UP000078492 UP000075886 UP000075809 UP000075920 UP000078542 UP000007062 UP000053097 UP000000311 UP000078541 UP000030742 UP000019118 UP000279307 UP000092462
UP000192221 UP000092444 UP000092443 UP000030765 UP000092445 UP000007798 UP000078200 UP000092460 UP000037069 UP000007801 UP000268350 UP000069940 UP000249989 UP000095301 UP000000304 UP000008744 UP000001819 UP000000803 UP000001292 UP000092553 UP000001070 UP000095300 UP000009192 UP000002320 UP000008820 UP000008711 UP000183832 UP000002282 UP000008792 UP000075884 UP000007266 UP000075880 UP000078492 UP000075886 UP000075809 UP000075920 UP000078542 UP000007062 UP000053097 UP000000311 UP000078541 UP000030742 UP000019118 UP000279307 UP000092462
Pfam
Interpro
IPR036259
MFS_trans_sf
+ More
IPR002666 Folate_carrier
IPR020846 MFS_dom
IPR036028 SH3-like_dom_sf
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR000261 EH_dom
IPR001452 SH3_domain
IPR027005 GlyclTrfase_39-like
IPR003342 Glyco_trans_39/83
IPR036300 MIR_dom_sf
IPR016093 MIR_motif
IPR032421 PMT_4TMC
IPR036186 Serpin_sf
IPR023796 Serpin_dom
IPR042178 Serpin_sf_1
IPR023795 Serpin_CS
IPR042185 Serpin_sf_2
IPR002666 Folate_carrier
IPR020846 MFS_dom
IPR036028 SH3-like_dom_sf
IPR002048 EF_hand_dom
IPR011992 EF-hand-dom_pair
IPR018247 EF_Hand_1_Ca_BS
IPR000261 EH_dom
IPR001452 SH3_domain
IPR027005 GlyclTrfase_39-like
IPR003342 Glyco_trans_39/83
IPR036300 MIR_dom_sf
IPR016093 MIR_motif
IPR032421 PMT_4TMC
IPR036186 Serpin_sf
IPR023796 Serpin_dom
IPR042178 Serpin_sf_1
IPR023795 Serpin_CS
IPR042185 Serpin_sf_2
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4K9B9
A0A437B9I2
A0A194RIX0
A0A212EXR6
A0A1E1WJ69
A0A194PR11
+ More
A0A1A9X432 A0A1W4V4M3 A0A1B0FNL7 A0A1A9YFY6 A0A084VJC1 A0A1B0AF25 B4NKW1 A0A1A9VU41 A0A1B0BH53 A0A0L0C7D6 B3LZG0 A0A023ESB1 A0A3B0KBK3 A0A182G3B7 A0A1I8N4B8 A0A1L8E8F6 B4QUM9 A0A1L8DE13 A0A1L8E8M5 B4GLC6 A0A1L8E8P8 Q294A4 Q9VGV5 B3LZF8 B4HIN5 A0A1L8EFX5 A0A0M3QYC5 A0A1L8EFP6 A0A0K8U209 A0A0A1XPW3 B4JGE1 A0A1L8EFP2 A0A1I8PS62 B4K5T8 B4JGE0 B0WKU6 J9EAM5 B3P1I7 A0A1J1IHM5 A0A034VQF7 B4PLE7 A0A1W4V4Y9 B4LZX8 Q9VGV6 A0A182N673 B4HIN6 B4LZX7 A0A1Q3FH43 B4PLE6 D2A676 A0A182JKS1 A0A195DKU9 W8C661 A0A182QBU7 A0A182NKA2 B3LZF9 A0A151X988 B4QUN0 B0WCL0 B4K5T9 B3P1I8 A0A336M170 B4GLC5 A0A1B0G7A7 A0A182WCD3 Q294A5 A0A195CXW0 Q7Q5N2 B4NKW0 A0A026WFU5 A0A0M4EGB4 A0A182QPN5 A0A336MEJ3 A0A0K8UQZ4 A0A182G1M5 E2A357 D6WJH6 A0A151JVH3 B3MNJ2 A0A1A9UGV0 A0A1Y1KHU6 U4UKF2 E9IXH9 A0A1B0C4Z9 A0A336LUE0 N6UCU0 E9IYY4 U4U6M1 A0A3L8DKQ6 A0A1B0DN88 A0A1A9XUE4 A0A1J1IHU2
A0A1A9X432 A0A1W4V4M3 A0A1B0FNL7 A0A1A9YFY6 A0A084VJC1 A0A1B0AF25 B4NKW1 A0A1A9VU41 A0A1B0BH53 A0A0L0C7D6 B3LZG0 A0A023ESB1 A0A3B0KBK3 A0A182G3B7 A0A1I8N4B8 A0A1L8E8F6 B4QUM9 A0A1L8DE13 A0A1L8E8M5 B4GLC6 A0A1L8E8P8 Q294A4 Q9VGV5 B3LZF8 B4HIN5 A0A1L8EFX5 A0A0M3QYC5 A0A1L8EFP6 A0A0K8U209 A0A0A1XPW3 B4JGE1 A0A1L8EFP2 A0A1I8PS62 B4K5T8 B4JGE0 B0WKU6 J9EAM5 B3P1I7 A0A1J1IHM5 A0A034VQF7 B4PLE7 A0A1W4V4Y9 B4LZX8 Q9VGV6 A0A182N673 B4HIN6 B4LZX7 A0A1Q3FH43 B4PLE6 D2A676 A0A182JKS1 A0A195DKU9 W8C661 A0A182QBU7 A0A182NKA2 B3LZF9 A0A151X988 B4QUN0 B0WCL0 B4K5T9 B3P1I8 A0A336M170 B4GLC5 A0A1B0G7A7 A0A182WCD3 Q294A5 A0A195CXW0 Q7Q5N2 B4NKW0 A0A026WFU5 A0A0M4EGB4 A0A182QPN5 A0A336MEJ3 A0A0K8UQZ4 A0A182G1M5 E2A357 D6WJH6 A0A151JVH3 B3MNJ2 A0A1A9UGV0 A0A1Y1KHU6 U4UKF2 E9IXH9 A0A1B0C4Z9 A0A336LUE0 N6UCU0 E9IYY4 U4U6M1 A0A3L8DKQ6 A0A1B0DN88 A0A1A9XUE4 A0A1J1IHU2
Ontologies
GO
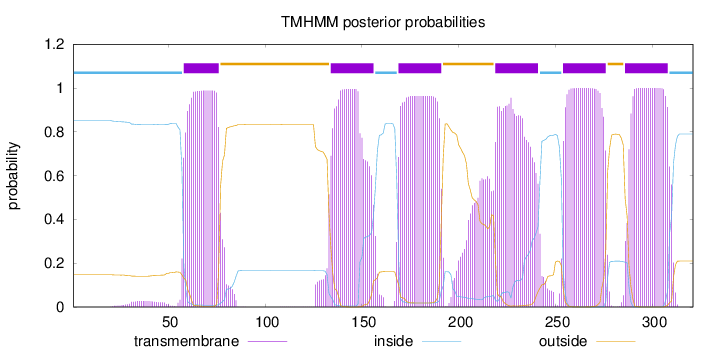
Topology
Length:
321
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
137.88482
Exp number, first 60 AAs:
3.21162
Total prob of N-in:
0.85284
inside
1 - 57
TMhelix
58 - 76
outside
77 - 133
TMhelix
134 - 156
inside
157 - 168
TMhelix
169 - 191
outside
192 - 218
TMhelix
219 - 241
inside
242 - 253
TMhelix
254 - 276
outside
277 - 285
TMhelix
286 - 308
inside
309 - 321
Population Genetic Test Statistics
Pi
252.573906
Theta
146.167759
Tajima's D
2.076348
CLR
17.505496
CSRT
0.890355482225889
Interpretation
Uncertain
