Gene
KWMTBOMO09217 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003404
Annotation
PREDICTED:_protein_takeout-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 2.259
Sequence
CDS
ATGATTTCTGTTCAAAATTTTTTGTTACCATCCTTTCTCGTATTATGTGTGGTCGGGAGCGGTCAATCGAAGAAACAGATCCCCGGCTACATACAAATTTGCAAACATGATCCGAATACAATCAGCGAATGCGTGCAGAAATCTATTGAGGCGCTAAAACCTAAATTAGCCGAAGGCATCCCCGAACTGGACGTTCCACCGTTAGAACCCTTCATCATTCCCGAGGTGGTGGCTTCCAGTAAAGATCGTGTGCCGCTTCAAGCTACTGGAAAAAATATCAAGGTCACGGGGGCTGGTGATTTTCATATTAATAAATTGAATGTGGATCTCGATAGTTTGATATTGAAAGCCAAGGTTAATTTCCCGAAGTTGCACTTCGACGGATTATACAAGTTGGACACTCAGATCTTGGTCGTGCCCATCAAGGGAGAGGGAAATCTTCTTGCTGACGTCGTGAACTGCGACGCCGAACTCGTGTTCAAAGGAGGGAAGTACCAGAAAGACGGTCTCGAATTCATTAAGTTCAATAGTCTTGATACTAAGATCGGAATCAAAGATTATAACGTTAAATTTGATGGATTGTTCAATAACGACAAAGTTCTCGGTGATGCAGCCAATGACGCAATCAATCAGAACAAGGGAGAGTTCTTCAATACGTTCAAGCCGTTCCTGGAGGCGGCTGTTTCAAAACTGCTCCTAAATATATCCAACAAAGTGGTCGACGACCTCCCATTCAACGAACTACTGCCCAAACCTTAA
Protein
MISVQNFLLPSFLVLCVVGSGQSKKQIPGYIQICKHDPNTISECVQKSIEALKPKLAEGIPELDVPPLEPFIIPEVVASSKDRVPLQATGKNIKVTGAGDFHINKLNVDLDSLILKAKVNFPKLHFDGLYKLDTQILVVPIKGEGNLLADVVNCDAELVFKGGKYQKDGLEFIKFNSLDTKIGIKDYNVKFDGLFNNDKVLGDAANDAINQNKGEFFNTFKPFLEAAVSKLLLNISNKVVDDLPFNELLPKP
Summary
Uniprot
A0A2K8GME7
A0A1L2DC02
A0A2A4KAC4
A0A3S2L6Y9
A0A3G1KKW4
A0A0K8TUN8
+ More
I4DJR2 A0A194RK02 A0A194PQZ7 D6WKU9 A0A2K8JM92 R4FK69 A0A0A9YGH0 A0A0K8TFW6 A0A1B6L2D3 A0A023F9I0 A0A1B6G3Q6 A0A1B6H564 A0A067R736 A0A2J7QTE3 E0VZK1 R4UW95 A0A1W4WAR8 A0A1B6FK71 A0A1Y1MXT2 A0A3Q0IZD1 A0A0T6B9L6 A0A1B6DN66 E0VZK3 D6WKU8 J3JYE7 A0A0A9ZFB0 B3P0X5 B4PR49 A0A0M5J2G7 A0A1W4VUQ4 A0A232FHL2 A0A146M7Z8 D6WLR1 B4HLE7 K7IUP9 A0A0C9QD95 Q9VEZ6 C1C588 B3MXB0 B7ZWP8 D6WKV0 A0A0C9QKV1 E0VZJ6 A0A087ZVB6 A0A2K8JLT4 A0A0M8ZVD0 A0A146L7U4 A0A0A9YPX6 T1HEN4 A0A182TCR6 A0A126GUV0 A0A1L8DQ60 B4JGA5 A0A1L8DQC1 A0A212EL67 A0A1Y1JV16 A0A0J7KZU9 A0A194RKA0 T1HLJ9 J3JWP5 A0A182V0Y1 A0A2C9GRK3 A0A182XBE1 A0A182KLU2 F5HMA4 A0A336MQ15 A0A182M548 A0A2J7QTG3 A0A084VHJ4 A0A1W4WAQ0 A0A3B0KC00 B4NL19 A0A1I8P3D9 A0A182VYY6 A0A3S2NS13 E2AH15 A0A1Y1NAE1 A0A182RPV6 B4LY18 I4DK41 A0A194PJR0 A0A2A4K9B6 B4GML5 Q294U4 K7IUP7 A0A0A9YBA1 A0A0C9RAB6 J9JT51 A0A3G1KL07 A0A182PKI0 A0A0K8SJR3 T1P8J1
I4DJR2 A0A194RK02 A0A194PQZ7 D6WKU9 A0A2K8JM92 R4FK69 A0A0A9YGH0 A0A0K8TFW6 A0A1B6L2D3 A0A023F9I0 A0A1B6G3Q6 A0A1B6H564 A0A067R736 A0A2J7QTE3 E0VZK1 R4UW95 A0A1W4WAR8 A0A1B6FK71 A0A1Y1MXT2 A0A3Q0IZD1 A0A0T6B9L6 A0A1B6DN66 E0VZK3 D6WKU8 J3JYE7 A0A0A9ZFB0 B3P0X5 B4PR49 A0A0M5J2G7 A0A1W4VUQ4 A0A232FHL2 A0A146M7Z8 D6WLR1 B4HLE7 K7IUP9 A0A0C9QD95 Q9VEZ6 C1C588 B3MXB0 B7ZWP8 D6WKV0 A0A0C9QKV1 E0VZJ6 A0A087ZVB6 A0A2K8JLT4 A0A0M8ZVD0 A0A146L7U4 A0A0A9YPX6 T1HEN4 A0A182TCR6 A0A126GUV0 A0A1L8DQ60 B4JGA5 A0A1L8DQC1 A0A212EL67 A0A1Y1JV16 A0A0J7KZU9 A0A194RKA0 T1HLJ9 J3JWP5 A0A182V0Y1 A0A2C9GRK3 A0A182XBE1 A0A182KLU2 F5HMA4 A0A336MQ15 A0A182M548 A0A2J7QTG3 A0A084VHJ4 A0A1W4WAQ0 A0A3B0KC00 B4NL19 A0A1I8P3D9 A0A182VYY6 A0A3S2NS13 E2AH15 A0A1Y1NAE1 A0A182RPV6 B4LY18 I4DK41 A0A194PJR0 A0A2A4K9B6 B4GML5 Q294U4 K7IUP7 A0A0A9YBA1 A0A0C9RAB6 J9JT51 A0A3G1KL07 A0A182PKI0 A0A0K8SJR3 T1P8J1
Pubmed
29380573
28973484
26017144
22651552
26354079
18362917
+ More
19820115 25401762 26823975 25474469 24845553 20566863 28004739 22516182 23537049 17994087 17550304 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22118469 20966253 12364791 14747013 17210077 24438588 20798317 15632085
19820115 25401762 26823975 25474469 24845553 20566863 28004739 22516182 23537049 17994087 17550304 28648823 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22118469 20966253 12364791 14747013 17210077 24438588 20798317 15632085
EMBL
KY681053
ART46240.1
KU981054
ANA78435.1
NWSH01000012
PCG80864.1
+ More
RSAL01000112 RVE47052.1 MF196301 ATU07283.1 GCVX01000237 JAI17993.1 AK401530 BAM18152.1 KQ460106 KPJ17764.1 KQ459602 KPI93555.1 KQ971343 EFA04020.1 KY031010 ATU82761.1 ACPB03005485 GAHY01001998 JAA75512.1 GBHO01013401 JAG30203.1 GBRD01001407 GDHC01011139 JAG64414.1 JAQ07490.1 GEBQ01022161 JAT17816.1 GBBI01001139 JAC17573.1 GECZ01012691 GECZ01005379 JAS57078.1 JAS64390.1 GECU01037898 GECU01032168 GECU01024220 GECU01013025 JAS69808.1 JAS75538.1 JAS83486.1 JAS94681.1 KK852901 KDR14111.1 NEVH01011194 PNF31861.1 DS235853 EEB18807.1 KC740702 AGM32526.1 GECZ01019184 JAS50585.1 GEZM01017697 GEZM01017695 JAV90492.1 LJIG01009028 KRT83899.1 GEDC01010201 JAS27097.1 EEB18809.1 EFA03557.2 APGK01037639 BT128273 KB740948 KB632364 AEE63233.1 ENN77412.1 ERL93584.1 GBHO01001511 JAG42093.1 CH954181 EDV49094.1 CM000160 EDW97371.1 CP012526 ALC47748.1 NNAY01000182 OXU30234.1 GDHC01002741 JAQ15888.1 EFA03416.2 CH480815 EDW41967.1 AAZX01004562 GBYB01012368 JAG82135.1 AE014297 AAF55266.1 AGB96002.1 BT082017 ACO72854.1 CH902629 EDV35333.1 BT050590 ACJ49255.1 EFA03556.1 GBYB01001192 JAG70959.1 EEB18802.1 KY031008 ATU82759.1 KQ435830 KOX71717.1 GDHC01014964 JAQ03665.1 GBHO01010476 JAG33128.1 ACPB03005486 ALI30574.1 GFDF01005553 JAV08531.1 CH916369 EDV92574.1 GFDF01005554 JAV08530.1 AGBW02014114 OWR42240.1 GEZM01102987 JAV51640.1 LBMM01001712 KMQ95853.1 KPJ17765.1 ACPB03014059 APGK01037640 BT127663 AEE62625.1 ENN77414.1 APCN01002054 AAAB01008987 EGK97426.1 UFQT01000450 UFQT01001404 SSX24488.1 SSX30387.1 AXCM01009717 PNF31863.1 ATLV01013177 KE524842 KFB37438.1 OUUW01000008 SPP83679.1 CH964272 EDW84222.1 RVE47051.1 GL439444 EFN67287.1 GEZM01013444 JAV92547.1 CH940650 EDW66884.1 AK401659 BAM18281.1 KPI93557.1 PCG80865.1 CH479185 EDW38089.1 CM000070 EAL28870.1 GBHO01016804 JAG26800.1 GBYB01005200 JAG74967.1 ABLF02029366 ABLF02029369 MF196308 ATU07290.1 GBRD01012415 JAG53409.1 KA644962 AFP59591.1
RSAL01000112 RVE47052.1 MF196301 ATU07283.1 GCVX01000237 JAI17993.1 AK401530 BAM18152.1 KQ460106 KPJ17764.1 KQ459602 KPI93555.1 KQ971343 EFA04020.1 KY031010 ATU82761.1 ACPB03005485 GAHY01001998 JAA75512.1 GBHO01013401 JAG30203.1 GBRD01001407 GDHC01011139 JAG64414.1 JAQ07490.1 GEBQ01022161 JAT17816.1 GBBI01001139 JAC17573.1 GECZ01012691 GECZ01005379 JAS57078.1 JAS64390.1 GECU01037898 GECU01032168 GECU01024220 GECU01013025 JAS69808.1 JAS75538.1 JAS83486.1 JAS94681.1 KK852901 KDR14111.1 NEVH01011194 PNF31861.1 DS235853 EEB18807.1 KC740702 AGM32526.1 GECZ01019184 JAS50585.1 GEZM01017697 GEZM01017695 JAV90492.1 LJIG01009028 KRT83899.1 GEDC01010201 JAS27097.1 EEB18809.1 EFA03557.2 APGK01037639 BT128273 KB740948 KB632364 AEE63233.1 ENN77412.1 ERL93584.1 GBHO01001511 JAG42093.1 CH954181 EDV49094.1 CM000160 EDW97371.1 CP012526 ALC47748.1 NNAY01000182 OXU30234.1 GDHC01002741 JAQ15888.1 EFA03416.2 CH480815 EDW41967.1 AAZX01004562 GBYB01012368 JAG82135.1 AE014297 AAF55266.1 AGB96002.1 BT082017 ACO72854.1 CH902629 EDV35333.1 BT050590 ACJ49255.1 EFA03556.1 GBYB01001192 JAG70959.1 EEB18802.1 KY031008 ATU82759.1 KQ435830 KOX71717.1 GDHC01014964 JAQ03665.1 GBHO01010476 JAG33128.1 ACPB03005486 ALI30574.1 GFDF01005553 JAV08531.1 CH916369 EDV92574.1 GFDF01005554 JAV08530.1 AGBW02014114 OWR42240.1 GEZM01102987 JAV51640.1 LBMM01001712 KMQ95853.1 KPJ17765.1 ACPB03014059 APGK01037640 BT127663 AEE62625.1 ENN77414.1 APCN01002054 AAAB01008987 EGK97426.1 UFQT01000450 UFQT01001404 SSX24488.1 SSX30387.1 AXCM01009717 PNF31863.1 ATLV01013177 KE524842 KFB37438.1 OUUW01000008 SPP83679.1 CH964272 EDW84222.1 RVE47051.1 GL439444 EFN67287.1 GEZM01013444 JAV92547.1 CH940650 EDW66884.1 AK401659 BAM18281.1 KPI93557.1 PCG80865.1 CH479185 EDW38089.1 CM000070 EAL28870.1 GBHO01016804 JAG26800.1 GBYB01005200 JAG74967.1 ABLF02029366 ABLF02029369 MF196308 ATU07290.1 GBRD01012415 JAG53409.1 KA644962 AFP59591.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000007266
UP000015103
+ More
UP000027135 UP000235965 UP000009046 UP000192223 UP000079169 UP000019118 UP000030742 UP000008711 UP000002282 UP000092553 UP000192221 UP000215335 UP000001292 UP000002358 UP000000803 UP000007801 UP000005203 UP000053105 UP000075901 UP000001070 UP000007151 UP000036403 UP000075903 UP000075840 UP000076407 UP000075882 UP000007062 UP000075883 UP000030765 UP000268350 UP000007798 UP000095300 UP000075920 UP000000311 UP000075900 UP000008792 UP000008744 UP000001819 UP000007819 UP000075885
UP000027135 UP000235965 UP000009046 UP000192223 UP000079169 UP000019118 UP000030742 UP000008711 UP000002282 UP000092553 UP000192221 UP000215335 UP000001292 UP000002358 UP000000803 UP000007801 UP000005203 UP000053105 UP000075901 UP000001070 UP000007151 UP000036403 UP000075903 UP000075840 UP000076407 UP000075882 UP000007062 UP000075883 UP000030765 UP000268350 UP000007798 UP000095300 UP000075920 UP000000311 UP000075900 UP000008792 UP000008744 UP000001819 UP000007819 UP000075885
PRIDE
Pfam
PF06585 JHBP
Interpro
SUPFAM
SSF55394
SSF55394
Gene 3D
ProteinModelPortal
A0A2K8GME7
A0A1L2DC02
A0A2A4KAC4
A0A3S2L6Y9
A0A3G1KKW4
A0A0K8TUN8
+ More
I4DJR2 A0A194RK02 A0A194PQZ7 D6WKU9 A0A2K8JM92 R4FK69 A0A0A9YGH0 A0A0K8TFW6 A0A1B6L2D3 A0A023F9I0 A0A1B6G3Q6 A0A1B6H564 A0A067R736 A0A2J7QTE3 E0VZK1 R4UW95 A0A1W4WAR8 A0A1B6FK71 A0A1Y1MXT2 A0A3Q0IZD1 A0A0T6B9L6 A0A1B6DN66 E0VZK3 D6WKU8 J3JYE7 A0A0A9ZFB0 B3P0X5 B4PR49 A0A0M5J2G7 A0A1W4VUQ4 A0A232FHL2 A0A146M7Z8 D6WLR1 B4HLE7 K7IUP9 A0A0C9QD95 Q9VEZ6 C1C588 B3MXB0 B7ZWP8 D6WKV0 A0A0C9QKV1 E0VZJ6 A0A087ZVB6 A0A2K8JLT4 A0A0M8ZVD0 A0A146L7U4 A0A0A9YPX6 T1HEN4 A0A182TCR6 A0A126GUV0 A0A1L8DQ60 B4JGA5 A0A1L8DQC1 A0A212EL67 A0A1Y1JV16 A0A0J7KZU9 A0A194RKA0 T1HLJ9 J3JWP5 A0A182V0Y1 A0A2C9GRK3 A0A182XBE1 A0A182KLU2 F5HMA4 A0A336MQ15 A0A182M548 A0A2J7QTG3 A0A084VHJ4 A0A1W4WAQ0 A0A3B0KC00 B4NL19 A0A1I8P3D9 A0A182VYY6 A0A3S2NS13 E2AH15 A0A1Y1NAE1 A0A182RPV6 B4LY18 I4DK41 A0A194PJR0 A0A2A4K9B6 B4GML5 Q294U4 K7IUP7 A0A0A9YBA1 A0A0C9RAB6 J9JT51 A0A3G1KL07 A0A182PKI0 A0A0K8SJR3 T1P8J1
I4DJR2 A0A194RK02 A0A194PQZ7 D6WKU9 A0A2K8JM92 R4FK69 A0A0A9YGH0 A0A0K8TFW6 A0A1B6L2D3 A0A023F9I0 A0A1B6G3Q6 A0A1B6H564 A0A067R736 A0A2J7QTE3 E0VZK1 R4UW95 A0A1W4WAR8 A0A1B6FK71 A0A1Y1MXT2 A0A3Q0IZD1 A0A0T6B9L6 A0A1B6DN66 E0VZK3 D6WKU8 J3JYE7 A0A0A9ZFB0 B3P0X5 B4PR49 A0A0M5J2G7 A0A1W4VUQ4 A0A232FHL2 A0A146M7Z8 D6WLR1 B4HLE7 K7IUP9 A0A0C9QD95 Q9VEZ6 C1C588 B3MXB0 B7ZWP8 D6WKV0 A0A0C9QKV1 E0VZJ6 A0A087ZVB6 A0A2K8JLT4 A0A0M8ZVD0 A0A146L7U4 A0A0A9YPX6 T1HEN4 A0A182TCR6 A0A126GUV0 A0A1L8DQ60 B4JGA5 A0A1L8DQC1 A0A212EL67 A0A1Y1JV16 A0A0J7KZU9 A0A194RKA0 T1HLJ9 J3JWP5 A0A182V0Y1 A0A2C9GRK3 A0A182XBE1 A0A182KLU2 F5HMA4 A0A336MQ15 A0A182M548 A0A2J7QTG3 A0A084VHJ4 A0A1W4WAQ0 A0A3B0KC00 B4NL19 A0A1I8P3D9 A0A182VYY6 A0A3S2NS13 E2AH15 A0A1Y1NAE1 A0A182RPV6 B4LY18 I4DK41 A0A194PJR0 A0A2A4K9B6 B4GML5 Q294U4 K7IUP7 A0A0A9YBA1 A0A0C9RAB6 J9JT51 A0A3G1KL07 A0A182PKI0 A0A0K8SJR3 T1P8J1
PDB
4G0S
E-value=4.7676e-06,
Score=118
Ontologies
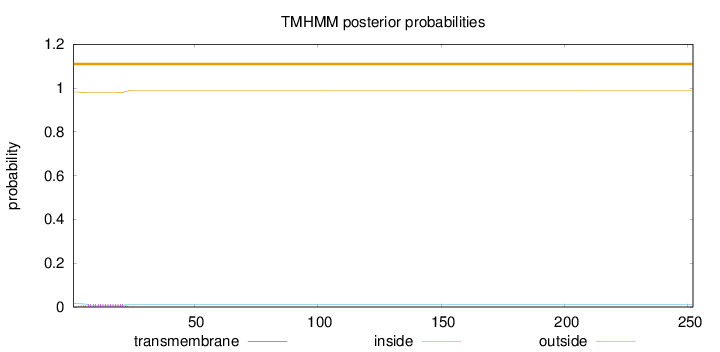
Topology
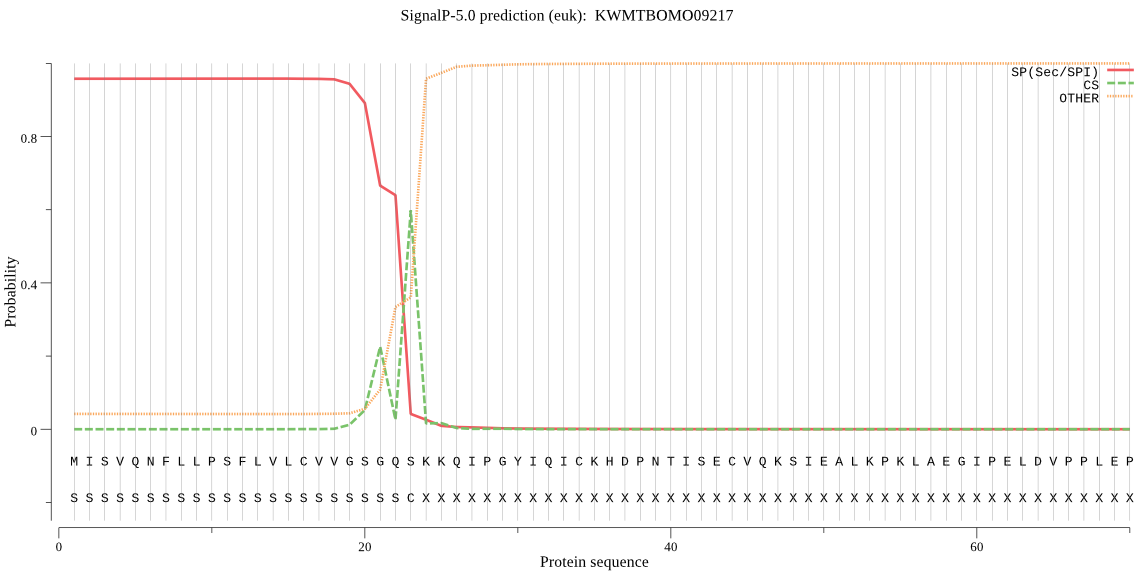
SignalP
Position: 1 - 23,
Likelihood: 0.958039
Length:
252
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24079
Exp number, first 60 AAs:
0.24058
Total prob of N-in:
0.01835
outside
1 - 252
Population Genetic Test Statistics
Pi
218.770783
Theta
205.799753
Tajima's D
0.098345
CLR
211.789351
CSRT
0.394330283485826
Interpretation
Uncertain
