Pre Gene Modal
BGIBMGA003402
Annotation
archease_[Bombyx_mori]
Full name
Protein archease-like
Location in the cell
Cytoplasmic Reliability : 2.296
Sequence
CDS
ATGGATGAAGAATTAGGCGGTATTTCCGAAGAATATTATGATATACCTCCTGTTAAATATGAATATTTGGACCATACTGCTGATGTTCAATTACACGCATGGGGCGACAGTCTTAAAGAAGCCTTCGAGCAGTGCGGGATGGCGATGTTCGGTTACATGACCGAGCTTGAGTACGTCCAGATCAAGGATGTTCACACCATCGAAGCGAGCGCTGACGATCTCATGGGATTACTTTACCACTTCCTCGACGAACTGTTGTTTTTATTCTCCTACGAACCATTTTTGATATGCAAGAAGTTAGTCATAACGGAGTTTAACACGCAAGAGTTCAAGATAGTGTGTAGGTGTTACGGCGAGGAGTTCGTAATTGGCAAACATCCCCAAGGCACAGAAGTCAAAGCTATAACTTACTCGGCCATGCAGATCATTGACGATCCAAAGGACAATAAGTACGAAGTGTTTGTTATCATTGACATATAA
Protein
MDEELGGISEEYYDIPPVKYEYLDHTADVQLHAWGDSLKEAFEQCGMAMFGYMTELEYVQIKDVHTIEASADDLMGLLYHFLDELLFLFSYEPFLICKKLVITEFNTQEFKIVCRCYGEEFVIGKHPQGTEVKAITYSAMQIIDDPKDNKYEVFVIIDI
Summary
Description
Component of the tRNA-splicing ligase complex required to facilitate the enzymatic turnover of catalytic subunit RtcB (CG9987).
Similarity
Belongs to the archease family.
Keywords
Calcium
Complete proteome
Metal-binding
Reference proteome
tRNA processing
Feature
chain Protein archease-like
Uniprot
Q1HPX5
A0A2A4KB50
A0A2H1WEF7
A0A212ENQ2
A0A437B8Q5
S4NXC6
+ More
I4DKV1 A0A0L7L6M8 A0A1E1W737 A0A194RJZ7 D6WKY5 A0A1Y1MJT2 A0A1Y1MHT1 B3M0S8 A0A1B0GF44 A0A1A9W2C5 A0A034WQB4 A0A0L0C931 A0A0M4EH88 A0A1B0BBM6 A0A1I8PWK9 B4N8K3 A0A1A9YK03 A0A1W4X5H0 A0A1B0A0D5 J3JY63 A0A0K8WJ82 A0A1L8EDU5 A0A1A9UYH2 A0A1I8NF25 N6ULH7 B4PQW7 A0A0A1WLA8 B4IV27 B3NYV8 W8BNK3 B4LZ50 A0A3B0KJG7 Q295K3 B4GDR4 A0A1W4VKS5 B4KBJ6 B4I3E4 B4R0E1 B4JGW3 H0RNK8 Q9VD92 A0A026VU70 A0A182GMJ5 A0A151IVF1 A0A0K8TP52 A0A182RPW2 F4X7V9 E9IQV9 Q16KZ9 A0A1S4FX94 W5JKK5 A0A0T6BGV9 W8CCD9 A0A182XVT3 A0A2M4AT22 A0A182F6S5 A0A2M4ATM5 A0A2M4ATQ2 A0A195FXG6 A0A182J032 A0A182KAH2 U5EWL4 A0A182VYX9 A0A084VHK8 A0A182VN96 A0A182TDK1 A0A182XBE7 A0A182SR17 Q7PYJ8 A0A182Q0E3 A0A1Y1MJY4 A0A182NS90 A0A182MPK0 A0A195BTF4 A0A158NUG1 E2BMP9 A0A182PKH5 A0A1Y1MMR7 A0A182I088 B0XLR6 A0A182KLW2 A0A195BZ62 A0A1B6KSH9 E2AH10 A0A0L7RGN7 K7IUQ3 A0A0C9R2I9 A0A023EGZ0 A0A1L8DB54 A0A0N0BEI8 A0A1B6GXS3 A0A131Y5W9 A0A1Q3FCL4 A0A1B0CVW3
I4DKV1 A0A0L7L6M8 A0A1E1W737 A0A194RJZ7 D6WKY5 A0A1Y1MJT2 A0A1Y1MHT1 B3M0S8 A0A1B0GF44 A0A1A9W2C5 A0A034WQB4 A0A0L0C931 A0A0M4EH88 A0A1B0BBM6 A0A1I8PWK9 B4N8K3 A0A1A9YK03 A0A1W4X5H0 A0A1B0A0D5 J3JY63 A0A0K8WJ82 A0A1L8EDU5 A0A1A9UYH2 A0A1I8NF25 N6ULH7 B4PQW7 A0A0A1WLA8 B4IV27 B3NYV8 W8BNK3 B4LZ50 A0A3B0KJG7 Q295K3 B4GDR4 A0A1W4VKS5 B4KBJ6 B4I3E4 B4R0E1 B4JGW3 H0RNK8 Q9VD92 A0A026VU70 A0A182GMJ5 A0A151IVF1 A0A0K8TP52 A0A182RPW2 F4X7V9 E9IQV9 Q16KZ9 A0A1S4FX94 W5JKK5 A0A0T6BGV9 W8CCD9 A0A182XVT3 A0A2M4AT22 A0A182F6S5 A0A2M4ATM5 A0A2M4ATQ2 A0A195FXG6 A0A182J032 A0A182KAH2 U5EWL4 A0A182VYX9 A0A084VHK8 A0A182VN96 A0A182TDK1 A0A182XBE7 A0A182SR17 Q7PYJ8 A0A182Q0E3 A0A1Y1MJY4 A0A182NS90 A0A182MPK0 A0A195BTF4 A0A158NUG1 E2BMP9 A0A182PKH5 A0A1Y1MMR7 A0A182I088 B0XLR6 A0A182KLW2 A0A195BZ62 A0A1B6KSH9 E2AH10 A0A0L7RGN7 K7IUQ3 A0A0C9R2I9 A0A023EGZ0 A0A1L8DB54 A0A0N0BEI8 A0A1B6GXS3 A0A131Y5W9 A0A1Q3FCL4 A0A1B0CVW3
Pubmed
19121390
22118469
23622113
22651552
26354079
26227816
+ More
18362917 19820115 28004739 17994087 25348373 26108605 22516182 25315136 23537049 17550304 25830018 24495485 15632085 10731132 12537572 24508170 30249741 26483478 26369729 21719571 21282665 17510324 20920257 23761445 25244985 24438588 12364791 14747013 17210077 21347285 20798317 20966253 20075255 24945155 29652888
18362917 19820115 28004739 17994087 25348373 26108605 22516182 25315136 23537049 17550304 25830018 24495485 15632085 10731132 12537572 24508170 30249741 26483478 26369729 21719571 21282665 17510324 20920257 23761445 25244985 24438588 12364791 14747013 17210077 21347285 20798317 20966253 20075255 24945155 29652888
EMBL
BABH01023786
DQ443277
ABF51366.1
NWSH01000012
PCG80892.1
ODYU01008115
+ More
SOQ51441.1 AGBW02013624 OWR43114.1 RSAL01000116 RVE46848.1 GAIX01009104 JAA83456.1 AK401919 KQ459602 BAM18541.1 KPI93550.1 JTDY01002633 KOB71050.1 GDQN01008393 JAT82661.1 KQ460106 KPJ17759.1 KQ971343 EFA04039.1 GEZM01030905 GEZM01030902 GEZM01030890 GEZM01030883 GEZM01030880 JAV85198.1 GEZM01030895 JAV85211.1 CH902617 EDV42093.1 CCAG010012761 GAKP01002425 JAC56527.1 JRES01000752 KNC28752.1 CP012526 ALC45658.1 JXJN01011551 CH964232 EDW81454.1 BT128187 AEE63148.1 GDHF01001162 JAI51152.1 GFDG01001963 JAV16836.1 APGK01028352 KB740686 KB631930 ENN79557.1 ERL87314.1 CM000160 EDW96291.1 GBXI01014821 JAC99470.1 CH892596 EDX00241.1 CH954181 EDV48221.1 GAMC01003865 JAC02691.1 CH940650 EDW67057.1 OUUW01000008 SPP83898.1 CM000070 EAL28708.1 CH479182 EDW34575.1 CH933806 EDW13663.1 CH480821 EDW55308.1 CM000364 EDX12066.1 CH916369 EDV93741.1 BT132930 AEW12902.1 AE014297 BT003491 KK107899 QOIP01000006 EZA47282.1 RLU21557.1 JXUM01074847 KQ562855 KXJ74990.1 KQ980900 KYN11693.1 GDAI01001446 JAI16157.1 GL888900 EGI57445.1 GL764976 EFZ17043.1 CH477929 EAT34985.1 ADMH02001255 ETN63410.1 LJIG01000443 KRT86497.1 GAMC01003866 JAC02690.1 GGFK01010599 MBW43920.1 GGFK01010812 MBW44133.1 GGFK01010864 MBW44185.1 KQ981193 KYN45128.1 GANO01001364 JAB58507.1 ATLV01013177 KE524842 KFB37452.1 AAAB01008987 EAA01158.2 AXCN02000204 GEZM01030910 GEZM01030909 GEZM01030904 GEZM01030903 GEZM01030881 GEZM01030879 GEZM01030878 GEZM01030876 JAV85258.1 AXCM01013848 KQ976417 KYM89958.1 ADTU01026421 GL449285 EFN83034.1 GEZM01030896 GEZM01030894 GEZM01030886 GEZM01030885 JAV85236.1 APCN01002055 DS234676 EDS34731.1 KQ978501 KYM93615.1 GEBQ01025574 JAT14403.1 GL439444 EFN67282.1 KQ414598 KOC69856.1 AAZX01010814 GBYB01010314 JAG80081.1 GAPW01005509 JAC08089.1 GFDF01010504 JAV03580.1 KQ435830 KOX71722.1 GECZ01002572 JAS67197.1 GEFM01002350 GEGO01000804 JAP73446.1 JAR94600.1 GFDL01009755 JAV25290.1 AJWK01031344
SOQ51441.1 AGBW02013624 OWR43114.1 RSAL01000116 RVE46848.1 GAIX01009104 JAA83456.1 AK401919 KQ459602 BAM18541.1 KPI93550.1 JTDY01002633 KOB71050.1 GDQN01008393 JAT82661.1 KQ460106 KPJ17759.1 KQ971343 EFA04039.1 GEZM01030905 GEZM01030902 GEZM01030890 GEZM01030883 GEZM01030880 JAV85198.1 GEZM01030895 JAV85211.1 CH902617 EDV42093.1 CCAG010012761 GAKP01002425 JAC56527.1 JRES01000752 KNC28752.1 CP012526 ALC45658.1 JXJN01011551 CH964232 EDW81454.1 BT128187 AEE63148.1 GDHF01001162 JAI51152.1 GFDG01001963 JAV16836.1 APGK01028352 KB740686 KB631930 ENN79557.1 ERL87314.1 CM000160 EDW96291.1 GBXI01014821 JAC99470.1 CH892596 EDX00241.1 CH954181 EDV48221.1 GAMC01003865 JAC02691.1 CH940650 EDW67057.1 OUUW01000008 SPP83898.1 CM000070 EAL28708.1 CH479182 EDW34575.1 CH933806 EDW13663.1 CH480821 EDW55308.1 CM000364 EDX12066.1 CH916369 EDV93741.1 BT132930 AEW12902.1 AE014297 BT003491 KK107899 QOIP01000006 EZA47282.1 RLU21557.1 JXUM01074847 KQ562855 KXJ74990.1 KQ980900 KYN11693.1 GDAI01001446 JAI16157.1 GL888900 EGI57445.1 GL764976 EFZ17043.1 CH477929 EAT34985.1 ADMH02001255 ETN63410.1 LJIG01000443 KRT86497.1 GAMC01003866 JAC02690.1 GGFK01010599 MBW43920.1 GGFK01010812 MBW44133.1 GGFK01010864 MBW44185.1 KQ981193 KYN45128.1 GANO01001364 JAB58507.1 ATLV01013177 KE524842 KFB37452.1 AAAB01008987 EAA01158.2 AXCN02000204 GEZM01030910 GEZM01030909 GEZM01030904 GEZM01030903 GEZM01030881 GEZM01030879 GEZM01030878 GEZM01030876 JAV85258.1 AXCM01013848 KQ976417 KYM89958.1 ADTU01026421 GL449285 EFN83034.1 GEZM01030896 GEZM01030894 GEZM01030886 GEZM01030885 JAV85236.1 APCN01002055 DS234676 EDS34731.1 KQ978501 KYM93615.1 GEBQ01025574 JAT14403.1 GL439444 EFN67282.1 KQ414598 KOC69856.1 AAZX01010814 GBYB01010314 JAG80081.1 GAPW01005509 JAC08089.1 GFDF01010504 JAV03580.1 KQ435830 KOX71722.1 GECZ01002572 JAS67197.1 GEFM01002350 GEGO01000804 JAP73446.1 JAR94600.1 GFDL01009755 JAV25290.1 AJWK01031344
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000053240 UP000007266 UP000007801 UP000092444 UP000091820 UP000037069 UP000092553 UP000092460 UP000095300 UP000007798 UP000092443 UP000192223 UP000092445 UP000078200 UP000095301 UP000019118 UP000030742 UP000002282 UP000008711 UP000008792 UP000268350 UP000001819 UP000008744 UP000192221 UP000009192 UP000001292 UP000000304 UP000001070 UP000000803 UP000053097 UP000279307 UP000069940 UP000249989 UP000078492 UP000075900 UP000007755 UP000008820 UP000000673 UP000076408 UP000069272 UP000078541 UP000075880 UP000075881 UP000075920 UP000030765 UP000075903 UP000075902 UP000076407 UP000075901 UP000007062 UP000075886 UP000075884 UP000075883 UP000078540 UP000005205 UP000008237 UP000075885 UP000075840 UP000002320 UP000075882 UP000078542 UP000000311 UP000053825 UP000002358 UP000053105 UP000092461
UP000053240 UP000007266 UP000007801 UP000092444 UP000091820 UP000037069 UP000092553 UP000092460 UP000095300 UP000007798 UP000092443 UP000192223 UP000092445 UP000078200 UP000095301 UP000019118 UP000030742 UP000002282 UP000008711 UP000008792 UP000268350 UP000001819 UP000008744 UP000192221 UP000009192 UP000001292 UP000000304 UP000001070 UP000000803 UP000053097 UP000279307 UP000069940 UP000249989 UP000078492 UP000075900 UP000007755 UP000008820 UP000000673 UP000076408 UP000069272 UP000078541 UP000075880 UP000075881 UP000075920 UP000030765 UP000075903 UP000075902 UP000076407 UP000075901 UP000007062 UP000075886 UP000075884 UP000075883 UP000078540 UP000005205 UP000008237 UP000075885 UP000075840 UP000002320 UP000075882 UP000078542 UP000000311 UP000053825 UP000002358 UP000053105 UP000092461
PRIDE
Pfam
PF01951 Archease
SUPFAM
SSF69819
SSF69819
Gene 3D
ProteinModelPortal
Q1HPX5
A0A2A4KB50
A0A2H1WEF7
A0A212ENQ2
A0A437B8Q5
S4NXC6
+ More
I4DKV1 A0A0L7L6M8 A0A1E1W737 A0A194RJZ7 D6WKY5 A0A1Y1MJT2 A0A1Y1MHT1 B3M0S8 A0A1B0GF44 A0A1A9W2C5 A0A034WQB4 A0A0L0C931 A0A0M4EH88 A0A1B0BBM6 A0A1I8PWK9 B4N8K3 A0A1A9YK03 A0A1W4X5H0 A0A1B0A0D5 J3JY63 A0A0K8WJ82 A0A1L8EDU5 A0A1A9UYH2 A0A1I8NF25 N6ULH7 B4PQW7 A0A0A1WLA8 B4IV27 B3NYV8 W8BNK3 B4LZ50 A0A3B0KJG7 Q295K3 B4GDR4 A0A1W4VKS5 B4KBJ6 B4I3E4 B4R0E1 B4JGW3 H0RNK8 Q9VD92 A0A026VU70 A0A182GMJ5 A0A151IVF1 A0A0K8TP52 A0A182RPW2 F4X7V9 E9IQV9 Q16KZ9 A0A1S4FX94 W5JKK5 A0A0T6BGV9 W8CCD9 A0A182XVT3 A0A2M4AT22 A0A182F6S5 A0A2M4ATM5 A0A2M4ATQ2 A0A195FXG6 A0A182J032 A0A182KAH2 U5EWL4 A0A182VYX9 A0A084VHK8 A0A182VN96 A0A182TDK1 A0A182XBE7 A0A182SR17 Q7PYJ8 A0A182Q0E3 A0A1Y1MJY4 A0A182NS90 A0A182MPK0 A0A195BTF4 A0A158NUG1 E2BMP9 A0A182PKH5 A0A1Y1MMR7 A0A182I088 B0XLR6 A0A182KLW2 A0A195BZ62 A0A1B6KSH9 E2AH10 A0A0L7RGN7 K7IUQ3 A0A0C9R2I9 A0A023EGZ0 A0A1L8DB54 A0A0N0BEI8 A0A1B6GXS3 A0A131Y5W9 A0A1Q3FCL4 A0A1B0CVW3
I4DKV1 A0A0L7L6M8 A0A1E1W737 A0A194RJZ7 D6WKY5 A0A1Y1MJT2 A0A1Y1MHT1 B3M0S8 A0A1B0GF44 A0A1A9W2C5 A0A034WQB4 A0A0L0C931 A0A0M4EH88 A0A1B0BBM6 A0A1I8PWK9 B4N8K3 A0A1A9YK03 A0A1W4X5H0 A0A1B0A0D5 J3JY63 A0A0K8WJ82 A0A1L8EDU5 A0A1A9UYH2 A0A1I8NF25 N6ULH7 B4PQW7 A0A0A1WLA8 B4IV27 B3NYV8 W8BNK3 B4LZ50 A0A3B0KJG7 Q295K3 B4GDR4 A0A1W4VKS5 B4KBJ6 B4I3E4 B4R0E1 B4JGW3 H0RNK8 Q9VD92 A0A026VU70 A0A182GMJ5 A0A151IVF1 A0A0K8TP52 A0A182RPW2 F4X7V9 E9IQV9 Q16KZ9 A0A1S4FX94 W5JKK5 A0A0T6BGV9 W8CCD9 A0A182XVT3 A0A2M4AT22 A0A182F6S5 A0A2M4ATM5 A0A2M4ATQ2 A0A195FXG6 A0A182J032 A0A182KAH2 U5EWL4 A0A182VYX9 A0A084VHK8 A0A182VN96 A0A182TDK1 A0A182XBE7 A0A182SR17 Q7PYJ8 A0A182Q0E3 A0A1Y1MJY4 A0A182NS90 A0A182MPK0 A0A195BTF4 A0A158NUG1 E2BMP9 A0A182PKH5 A0A1Y1MMR7 A0A182I088 B0XLR6 A0A182KLW2 A0A195BZ62 A0A1B6KSH9 E2AH10 A0A0L7RGN7 K7IUQ3 A0A0C9R2I9 A0A023EGZ0 A0A1L8DB54 A0A0N0BEI8 A0A1B6GXS3 A0A131Y5W9 A0A1Q3FCL4 A0A1B0CVW3
PDB
5YZ1
E-value=1.89395e-32,
Score=342
Ontologies
PANTHER
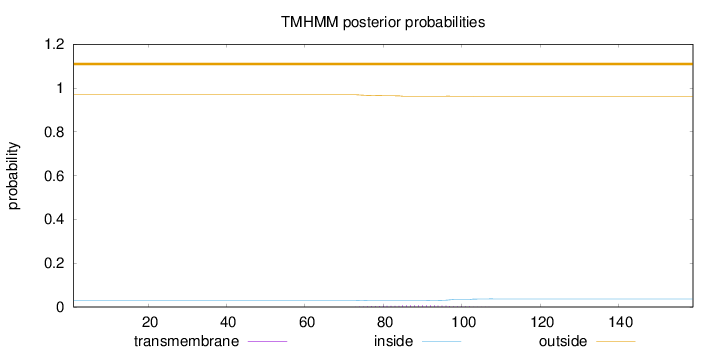
Topology
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15296
Exp number, first 60 AAs:
0.00039
Total prob of N-in:
0.03053
outside
1 - 159
Population Genetic Test Statistics
Pi
194.992777
Theta
144.268667
Tajima's D
-0.832873
CLR
0.359018
CSRT
0.164641767911604
Interpretation
Uncertain
