Gene
KWMTBOMO09206
Pre Gene Modal
BGIBMGA003349
Annotation
PREDICTED:_serine_protease_inhibitor_34_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.063
Sequence
CDS
ATGATACGCGGGTTGTGGTCAGCGAAGCGCAAGGCGCTACGACAGATCACAACCGTGGACACGATGCGCCGTAGTGGACGCGCGTTGTCCTGCGCGGCCGCAGGCCTGTTCTGCAACTTGTTATACGCGTGTATCAAGTTAACCGAGAGCACTGATTATGGAGTTCCGATAGAAATAAAGCTATGGGATACCGGACGCACACCTCTGGCACATATGGTGGATGTTACAAATGAGTTCGGCTTAAAAGTACTGGCGGAACATAATTTTCTGAACGAGAATAACATTGCGTTTTCGCCATACGGCTTAATGGGCATTCTGGTAGCCTTGTACGAAGGAGTCGATGGCGAGTCTTCATATCAACTGCAACGAGCGATACAGCTTCCGTGGAACCGAAAAATCATGAGAGTCGGGTTTCGAGACATTCACCGAACATTAAAAACTTATTTTGTTCCTGAAGAGGGTTTTTTGGCAGGTCTAGCATTAAATAATGAAAACGTTACATTCAATGAAAACTTTAAAAAAATATTACGTTTTTACGGCTTCGATTTGGATAATGATCAGCTTCCTGCCCTTCCTAATCAAACAAATAAGACTATGAATGAAACTACACCAAAATCCAGCACAATTGCTACGTCACAAGCAGATGGGGGTGATGTGTCTACAGGAACAACAGTTACAACAGAAAATACACGGGAAGAAACAACACCCGCGACATCTAGAGATACTACAGCAAATCCTAACGCCGAAACAATTAATACCATAGCAACAACGACTGCTATTCCATCTTCGGCACCGAATTTTGAAAGCACTACGGTTGGTATAAGTACCACAACACTTCAAGATATCATTAAACCACTAATATCTTCAACGGAAACTACAACACAAATCGTGACAACTACTAATTCTGTGACTACTTCTACGAACCCAACAGATTCAACGACTAACGAAAACTTATCGATCACCACGGCGAATCCTTCAAGTACGGATATTGACGAGGAAACGATAGCTCAGCAAACATCCCAAGCGACAGTGGCTTTAACACAAACCGAAAACAATGAAATCGATGGTACGGCGGCTGTGACTTCAAGTACAGTGGATTTGAACTTTACAGAAAGCACAACTTCTGTATCTACCACTGAAATACAGACCGATAGTGTCACATTTACAACATCAACAGACAAACCACTAGTTACTCTCTCTGATACGGCAACTAGTTCTGATATGACTACAGCTGCAAATGGTTCCTCGTTGGAAACACTTGAAAGGCGAAAGAAATCCATTGTTGATTTCATTTTCACGAACCCTCCTTACGTCGATGATTATCTAATGTATAGATCTTTTGATATACCTGCCGAGCCCCCAAAACCTACGTTCGATGAGCAAATGTTTTTGGCTAATGGATTAAAAAGCGTCCAGGTTACATACATGCATTACGACACTATATTAGAGCACGCATATCTGCCTCATCTAGAGGCTTCAGCTCTTCGGCTCCCATTAGACAGCGAACGATATTATCTCCTGGTTGTGTTGCCAGCCAGAGGGAGCGCCGCAGAATTAGGAAGATTGCTGGCACGTATGGCTCGTGAATCTGACTTGTCAGATATTTATGCGGCACTGCGTCCTAGGCGAGTCAAGGGAATAGTACCTAGTTTTACAGTAAAGGGCCACGTGACTCTCACTACAGATTTACAAAAGCTCGGAATACGAGATGTGTTTGAACCCAGACAGAGAGATTTCACACTAATGACTCGACAGTCTGGTGTTTACGTTCGAAATATTGAACAAGCCGTTTCTGTGGCCATTCGAAAATACCGCCCTGACGATCAAAAAAAAAATCGTAAGTCAACTTAA
Protein
MIRGLWSAKRKALRQITTVDTMRRSGRALSCAAAGLFCNLLYACIKLTESTDYGVPIEIKLWDTGRTPLAHMVDVTNEFGLKVLAEHNFLNENNIAFSPYGLMGILVALYEGVDGESSYQLQRAIQLPWNRKIMRVGFRDIHRTLKTYFVPEEGFLAGLALNNENVTFNENFKKILRFYGFDLDNDQLPALPNQTNKTMNETTPKSSTIATSQADGGDVSTGTTVTTENTREETTPATSRDTTANPNAETINTIATTTAIPSSAPNFESTTVGISTTTLQDIIKPLISSTETTTQIVTTTNSVTTSTNPTDSTTNENLSITTANPSSTDIDEETIAQQTSQATVALTQTENNEIDGTAAVTSSTVDLNFTESTTSVSTTEIQTDSVTFTTSTDKPLVTLSDTATSSDMTTAANGSSLETLERRKKSIVDFIFTNPPYVDDYLMYRSFDIPAEPPKPTFDEQMFLANGLKSVQVTYMHYDTILEHAYLPHLEASALRLPLDSERYYLLVVLPARGSAAELGRLLARMARESDLSDIYAALRPRRVKGIVPSFTVKGHVTLTTDLQKLGIRDVFEPRQRDFTLMTRQSGVYVRNIEQAVSVAIRKYRPDDQKKNRKST
Summary
Similarity
Belongs to the serpin family.
Proteomes
Pfam
PF00079 Serpin
Interpro
SUPFAM
SSF56574
SSF56574
Gene 3D
PDB
3NDA
E-value=5.02312e-05,
Score=113
Ontologies
GO
PANTHER
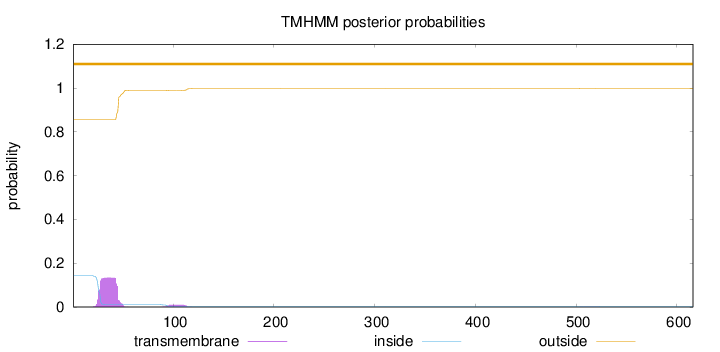
Topology
Length:
616
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.85643
Exp number, first 60 AAs:
2.65374
Total prob of N-in:
0.14355
outside
1 - 616
Population Genetic Test Statistics
Pi
239.850733
Theta
188.862741
Tajima's D
0.716634
CLR
0.725041
CSRT
0.584120793960302
Interpretation
Uncertain
