Pre Gene Modal
BGIBMGA003351
Annotation
minichromosome_maintenance_complex_component_7_[Bombyx_mori]
Full name
DNA replication licensing factor MCM7
+ More
DNA replication licensing factor Mcm7
DNA replication licensing factor mcm7-B
DNA replication licensing factor mcm7
DNA replication licensing factor mcm7-A
DNA replication licensing factor Mcm7
DNA replication licensing factor mcm7-B
DNA replication licensing factor mcm7
DNA replication licensing factor mcm7-A
Alternative Name
Minichromosome maintenance 7 protein
CDC47 homolog B
CDC47-2p
Minichromosome maintenance protein 7-B
CDC47 homolog
Minichromosome maintenance protein 7
CDC47 homolog A
CDC47p
Minichromosome maintenance protein 7-A
p90
CDC47 homolog B
CDC47-2p
Minichromosome maintenance protein 7-B
CDC47 homolog
Minichromosome maintenance protein 7
CDC47 homolog A
CDC47p
Minichromosome maintenance protein 7-A
p90
Location in the cell
Cytoplasmic Reliability : 2.308 Nuclear Reliability : 1.987
Sequence
CDS
ATGCGTGATTACACTGCAGATAAAGAATCTTTCAAGAACTTTTTCGTTGATTTTTGTCAAACTGATGATGAAGGAAAAAAGTATTTTAAATATGCTGAACAACTTACTAAAGTTGCACACAGAGAACAGATAGCATTTGAAGTTGACTTGGATGATTTACATGAGATGAATGAAGATTTGACTGAGGCAGTCAAACAAAATACCAGAAGATACACTAATATGGTGTCAGACGTGGTTTATGAAATGCTGCCTGATTACAAATTCAAAGAGGTAGTAGCAAAGGATTCCTTGGATGTATACATTGAGCACAGAATTATGTTGGAAGCCAGAAACCACAGGATTCCTGGAGAAATGAGGGATCCTAGGAACAGATACCCACCAGAGCTCATCAGGAGATTTGAAGTGTACTTCAAAGACTTGTCAACATCGAAAAGTGTACCTATCAGAGAAGTTAAGGCTGAACATATTGGGAAACTAGTTACTGTGCGAGGTACATATTATAGTATAGTCATGTTGCATATACGAATATAG
Protein
MRDYTADKESFKNFFVDFCQTDDEGKKYFKYAEQLTKVAHREQIAFEVDLDDLHEMNEDLTEAVKQNTRRYTNMVSDVVYEMLPDYKFKEVVAKDSLDVYIEHRIMLEARNHRIPGEMRDPRNRYPPELIRRFEVYFKDLSTSKSVPIREVKAEHIGKLVTVRGTYYSIVMLHIRI
Summary
Description
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. The existence of maternal and zygotic forms of mcm3 and mcm6 suggests that specific forms of mcm2-7 complexes may be used during different stages of development (By similarity).
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. The existence of maternal and zygotic forms of mcm3 and mcm6 suggests that specific forms of mcm2-7 complexes may be used during different stages of development.
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity.
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. The existence of maternal and zygotic forms of mcm3 and mcm6 suggests that specific forms of mcm2-7 complexes may be used during different stages of development (By similarity).
Acts as component of the mcm2-7 complex (mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. The existence of maternal and zygotic forms of mcm3 and mcm6 suggests that specific forms of mcm2-7 complexes may be used during different stages of development.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (Probable).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5. The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. The N-terminus is required for interaction with mmcm3, though this interaction may not be direct, and remains in a complex with mmcm3 throughout the cell cycle. Begins to associate with zmcm6 at the neurula stage (By similarity). Component of the replisome complex (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. The N-terminus is required for interaction with mmcm3, though this interaction may not be direct, and remains in a complex with mmcm3 throughout the cell cycle. Begins to associate with zmcm6 at the neurula stage. Component of the replisome complex (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5. The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. The N-terminus is required for interaction with mmcm3, though this interaction may not be direct, and remains in a complex with mmcm3 throughout the cell cycle. Begins to associate with zmcm6 at the neurula stage (By similarity). Component of the replisome complex (By similarity).
Component of the mcm2-7 complex (RLF-M). The complex forms a toroidal hexameric ring with the proposed subunit order mcm2-mcm6-mcm4-mcm7-mcm3-mcm5 (Probable). The heterodimer of mmcm3/mcm5 interacts with mcm4, mmcm6, mcm7 and weakly with mcm2. The N-terminus is required for interaction with mmcm3, though this interaction may not be direct, and remains in a complex with mmcm3 throughout the cell cycle. Begins to associate with zmcm6 at the neurula stage. Component of the replisome complex (By similarity).
Similarity
Belongs to the MCM family.
Keywords
ATP-binding
Cell cycle
Complete proteome
DNA replication
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Metal-binding
Zinc
Zinc-finger
Feature
chain DNA replication licensing factor Mcm7
Uniprot
Q60GE6
H9J1G4
A0A437AXE5
A0A2H1V398
A0A2A4K5P9
A0A194PQX7
+ More
A0A0L7KNN6 A0A194RP00 D6WK36 A0A2J7QTN3 A0A1B6CHG2 A0A090XB26 B4HK41 A0A131XVG5 A0A067R721 A0A131Z543 A0A1E1XG68 A0A2P8Y7I5 L7M6G7 A0A224YVW3 A0A1B6MDL9 A0A336MAI7 B4H9M2 A0A293N4B1 A0A1Z5KUK0 A0A1B6IDU9 B4QML9 B3NBM4 B4PFH7 Q9XYU0 B4KZG3 B7QHT6 A0A1W4VCI4 A0A0J9RS32 A0A3R7QWK7 A0A1B6GR93 Q29D32 E2AL70 A0A090XAY7 C3ZEX3 A0A3Q0ITS1 A0A3B0JBE8 B4LEP6 A0A1I8Q0B2 A0A0L0BMS2 A0A1J1IEB8 A0A0M4EGR2 A0A026WPZ1 N6TF82 W8BNL9 V5KXP1 B3M5X9 A0A0K8WEX4 A0A2P2I1N8 A0A151K3E9 A0A1L8H3C0 Q7ZXB1 B4MMD1 B4J3L8 A0A195FTX8 T1PBE8 W5KQY5 A0A1A9UP56 A0A0A1WYP5 A0A3B4B8T0 Q17ML5 A0A151X3K9 A0A1W2WB15 A0A151I2C1 A0A158NKZ4 A0A1B8Y939 Q6NX31 A0A0A9XGX9 F4WWT5 A0A1B0CLF2 E2BME7 A0A1Y1KVC9 A0A0J7KE46 A0A3B3RCA0 A0A1B0FC91 A0A0P4VTB0 A0A3Q0R3X1 A0A0C9QWH7 A0A1A9ZBZ2 A0A146XPY6 A0A423TWM4 A0A182GAT2 A0A1A9XQV9 A0A1B0ANR5 A0A1A9W3H0 B0VZP1 A0A084WDI5 Q91876 A0A087UTA1 A0A3B5KJS6 A0A2D0PT24 A0A3Q1BNY5 A0A3B4Z553 A0A2L2Y8F0
A0A0L7KNN6 A0A194RP00 D6WK36 A0A2J7QTN3 A0A1B6CHG2 A0A090XB26 B4HK41 A0A131XVG5 A0A067R721 A0A131Z543 A0A1E1XG68 A0A2P8Y7I5 L7M6G7 A0A224YVW3 A0A1B6MDL9 A0A336MAI7 B4H9M2 A0A293N4B1 A0A1Z5KUK0 A0A1B6IDU9 B4QML9 B3NBM4 B4PFH7 Q9XYU0 B4KZG3 B7QHT6 A0A1W4VCI4 A0A0J9RS32 A0A3R7QWK7 A0A1B6GR93 Q29D32 E2AL70 A0A090XAY7 C3ZEX3 A0A3Q0ITS1 A0A3B0JBE8 B4LEP6 A0A1I8Q0B2 A0A0L0BMS2 A0A1J1IEB8 A0A0M4EGR2 A0A026WPZ1 N6TF82 W8BNL9 V5KXP1 B3M5X9 A0A0K8WEX4 A0A2P2I1N8 A0A151K3E9 A0A1L8H3C0 Q7ZXB1 B4MMD1 B4J3L8 A0A195FTX8 T1PBE8 W5KQY5 A0A1A9UP56 A0A0A1WYP5 A0A3B4B8T0 Q17ML5 A0A151X3K9 A0A1W2WB15 A0A151I2C1 A0A158NKZ4 A0A1B8Y939 Q6NX31 A0A0A9XGX9 F4WWT5 A0A1B0CLF2 E2BME7 A0A1Y1KVC9 A0A0J7KE46 A0A3B3RCA0 A0A1B0FC91 A0A0P4VTB0 A0A3Q0R3X1 A0A0C9QWH7 A0A1A9ZBZ2 A0A146XPY6 A0A423TWM4 A0A182GAT2 A0A1A9XQV9 A0A1B0ANR5 A0A1A9W3H0 B0VZP1 A0A084WDI5 Q91876 A0A087UTA1 A0A3B5KJS6 A0A2D0PT24 A0A3Q1BNY5 A0A3B4Z553 A0A2L2Y8F0
EC Number
3.6.4.12
Pubmed
19121390
26354079
26227816
18362917
19820115
25970599
+ More
17994087 24845553 26830274 28503490 29403074 25576852 28797301 28528879 17550304 9795205 10023044 10731132 12537572 16798881 20122406 22936249 15632085 20798317 18563158 26108605 24508170 30249741 23537049 24495485 27762356 9214647 25315136 25329095 25830018 25463417 17510324 21347285 20431018 25401762 26823975 21719571 28004739 29240929 27129103 26483478 24438588 8816774 9214646 9512418 9851868 16369567 21551351 26561354
17994087 24845553 26830274 28503490 29403074 25576852 28797301 28528879 17550304 9795205 10023044 10731132 12537572 16798881 20122406 22936249 15632085 20798317 18563158 26108605 24508170 30249741 23537049 24495485 27762356 9214647 25315136 25329095 25830018 25463417 17510324 21347285 20431018 25401762 26823975 21719571 28004739 29240929 27129103 26483478 24438588 8816774 9214646 9512418 9851868 16369567 21551351 26561354
EMBL
AB177622
BAD61056.1
BABH01023773
BABH01023774
BABH01023775
RSAL01000296
+ More
RVE42822.1 ODYU01000468 SOQ35313.1 NWSH01000144 PCG79103.1 KQ459602 KPI93535.1 JTDY01007844 KOB64907.1 KQ460106 KPJ17746.1 KQ971342 EFA03620.1 NEVH01011194 PNF31951.1 GEDC01024392 JAS12906.1 GBIH01000593 JAC94117.1 CH480815 EDW40777.1 GEFM01004512 JAP71284.1 KK852901 KDR14096.1 GEDV01002991 JAP85566.1 GFAC01001097 JAT98091.1 PYGN01000833 PSN40226.1 GACK01005138 JAA59896.1 GFPF01007297 MAA18443.1 GEBQ01005944 JAT34033.1 UFQS01000305 UFQT01000305 SSX02684.1 SSX23058.1 CH479229 EDW36498.1 GFWV01022869 MAA47596.1 GFJQ02008203 JAV98766.1 GECU01022588 JAS85118.1 CM000363 EDX09773.1 CH954178 EDV50620.1 CM000159 EDW93103.1 AB010109 AF124743 AE014296 BT001526 CH933809 EDW17890.1 ABJB010049488 ABJB010084779 ABJB010367990 ABJB010443466 ABJB010659432 ABJB010997321 DS941638 EEC18408.1 CM002912 KMY98492.1 QCYY01001058 ROT80844.1 GECZ01004813 JAS64956.1 CH379070 EAL30582.1 GL440492 EFN65825.1 GBIH01000594 JAC94116.1 GG666612 EEN49279.1 OUUW01000002 SPP77352.1 CH940647 EDW69131.1 JRES01001641 KNC21307.1 CVRI01000047 CRK98114.1 CP012525 ALC43652.1 KK107152 QOIP01000007 EZA57154.1 RLU20693.1 APGK01006422 APGK01006423 APGK01040416 APGK01040417 KB740979 KB736759 ENN76428.1 ENN83143.1 GAMC01011579 JAB94976.1 KF471134 AHA42534.1 CH902618 EDV39669.1 GDHF01002715 JAI49599.1 IACF01002298 LAB67957.1 LKEY01042441 KYN50597.1 CM004470 OCT90585.1 U66710 BC045072 AAC60228.1 AAH45072.1 CH963847 EDW73276.1 CH916366 EDV96220.1 KQ981281 KYN43344.1 KA645984 AFP60613.1 GBXI01010103 JAD04189.1 CH477205 EAT47909.1 KQ982557 KYQ55026.1 KQ976546 KYM80958.1 ADTU01019058 KV460378 OCA19456.1 CR855766 BC067307 AAH67307.1 GBHO01025556 GDHC01009877 JAG18048.1 JAQ08752.1 GL888413 EGI61323.1 AJWK01017185 GL449192 EFN83138.1 GEZM01074886 JAV64421.1 LBMM01008881 KMQ88562.1 CCAG010011826 GDKW01000753 JAI55842.1 GBYB01000075 JAG69842.1 GCES01042175 JAR44148.1 ROT80847.1 JXUM01001904 KQ560130 KXJ84283.1 JXJN01000936 DS231815 EDS35133.1 ATLV01023022 KE525339 KFB48279.1 U51234 U44051 BC072932 KK121487 KFM80590.1 IAAA01009830 IAAA01009831 LAA04441.1
RVE42822.1 ODYU01000468 SOQ35313.1 NWSH01000144 PCG79103.1 KQ459602 KPI93535.1 JTDY01007844 KOB64907.1 KQ460106 KPJ17746.1 KQ971342 EFA03620.1 NEVH01011194 PNF31951.1 GEDC01024392 JAS12906.1 GBIH01000593 JAC94117.1 CH480815 EDW40777.1 GEFM01004512 JAP71284.1 KK852901 KDR14096.1 GEDV01002991 JAP85566.1 GFAC01001097 JAT98091.1 PYGN01000833 PSN40226.1 GACK01005138 JAA59896.1 GFPF01007297 MAA18443.1 GEBQ01005944 JAT34033.1 UFQS01000305 UFQT01000305 SSX02684.1 SSX23058.1 CH479229 EDW36498.1 GFWV01022869 MAA47596.1 GFJQ02008203 JAV98766.1 GECU01022588 JAS85118.1 CM000363 EDX09773.1 CH954178 EDV50620.1 CM000159 EDW93103.1 AB010109 AF124743 AE014296 BT001526 CH933809 EDW17890.1 ABJB010049488 ABJB010084779 ABJB010367990 ABJB010443466 ABJB010659432 ABJB010997321 DS941638 EEC18408.1 CM002912 KMY98492.1 QCYY01001058 ROT80844.1 GECZ01004813 JAS64956.1 CH379070 EAL30582.1 GL440492 EFN65825.1 GBIH01000594 JAC94116.1 GG666612 EEN49279.1 OUUW01000002 SPP77352.1 CH940647 EDW69131.1 JRES01001641 KNC21307.1 CVRI01000047 CRK98114.1 CP012525 ALC43652.1 KK107152 QOIP01000007 EZA57154.1 RLU20693.1 APGK01006422 APGK01006423 APGK01040416 APGK01040417 KB740979 KB736759 ENN76428.1 ENN83143.1 GAMC01011579 JAB94976.1 KF471134 AHA42534.1 CH902618 EDV39669.1 GDHF01002715 JAI49599.1 IACF01002298 LAB67957.1 LKEY01042441 KYN50597.1 CM004470 OCT90585.1 U66710 BC045072 AAC60228.1 AAH45072.1 CH963847 EDW73276.1 CH916366 EDV96220.1 KQ981281 KYN43344.1 KA645984 AFP60613.1 GBXI01010103 JAD04189.1 CH477205 EAT47909.1 KQ982557 KYQ55026.1 KQ976546 KYM80958.1 ADTU01019058 KV460378 OCA19456.1 CR855766 BC067307 AAH67307.1 GBHO01025556 GDHC01009877 JAG18048.1 JAQ08752.1 GL888413 EGI61323.1 AJWK01017185 GL449192 EFN83138.1 GEZM01074886 JAV64421.1 LBMM01008881 KMQ88562.1 CCAG010011826 GDKW01000753 JAI55842.1 GBYB01000075 JAG69842.1 GCES01042175 JAR44148.1 ROT80847.1 JXUM01001904 KQ560130 KXJ84283.1 JXJN01000936 DS231815 EDS35133.1 ATLV01023022 KE525339 KFB48279.1 U51234 U44051 BC072932 KK121487 KFM80590.1 IAAA01009830 IAAA01009831 LAA04441.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000037510
UP000053240
+ More
UP000007266 UP000235965 UP000001292 UP000027135 UP000245037 UP000008744 UP000000304 UP000008711 UP000002282 UP000000803 UP000009192 UP000001555 UP000192221 UP000283509 UP000001819 UP000000311 UP000001554 UP000079169 UP000268350 UP000008792 UP000095300 UP000037069 UP000183832 UP000092553 UP000053097 UP000279307 UP000019118 UP000007801 UP000078492 UP000186698 UP000007798 UP000001070 UP000078541 UP000095301 UP000018467 UP000078200 UP000261520 UP000008820 UP000075809 UP000078540 UP000005205 UP000008143 UP000007755 UP000092461 UP000008237 UP000036403 UP000261540 UP000092444 UP000261340 UP000092445 UP000069940 UP000249989 UP000092443 UP000092460 UP000091820 UP000002320 UP000030765 UP000054359 UP000005226 UP000221080 UP000257160 UP000261400
UP000007266 UP000235965 UP000001292 UP000027135 UP000245037 UP000008744 UP000000304 UP000008711 UP000002282 UP000000803 UP000009192 UP000001555 UP000192221 UP000283509 UP000001819 UP000000311 UP000001554 UP000079169 UP000268350 UP000008792 UP000095300 UP000037069 UP000183832 UP000092553 UP000053097 UP000279307 UP000019118 UP000007801 UP000078492 UP000186698 UP000007798 UP000001070 UP000078541 UP000095301 UP000018467 UP000078200 UP000261520 UP000008820 UP000075809 UP000078540 UP000005205 UP000008143 UP000007755 UP000092461 UP000008237 UP000036403 UP000261540 UP000092444 UP000261340 UP000092445 UP000069940 UP000249989 UP000092443 UP000092460 UP000091820 UP000002320 UP000030765 UP000054359 UP000005226 UP000221080 UP000257160 UP000261400
Interpro
ProteinModelPortal
Q60GE6
H9J1G4
A0A437AXE5
A0A2H1V398
A0A2A4K5P9
A0A194PQX7
+ More
A0A0L7KNN6 A0A194RP00 D6WK36 A0A2J7QTN3 A0A1B6CHG2 A0A090XB26 B4HK41 A0A131XVG5 A0A067R721 A0A131Z543 A0A1E1XG68 A0A2P8Y7I5 L7M6G7 A0A224YVW3 A0A1B6MDL9 A0A336MAI7 B4H9M2 A0A293N4B1 A0A1Z5KUK0 A0A1B6IDU9 B4QML9 B3NBM4 B4PFH7 Q9XYU0 B4KZG3 B7QHT6 A0A1W4VCI4 A0A0J9RS32 A0A3R7QWK7 A0A1B6GR93 Q29D32 E2AL70 A0A090XAY7 C3ZEX3 A0A3Q0ITS1 A0A3B0JBE8 B4LEP6 A0A1I8Q0B2 A0A0L0BMS2 A0A1J1IEB8 A0A0M4EGR2 A0A026WPZ1 N6TF82 W8BNL9 V5KXP1 B3M5X9 A0A0K8WEX4 A0A2P2I1N8 A0A151K3E9 A0A1L8H3C0 Q7ZXB1 B4MMD1 B4J3L8 A0A195FTX8 T1PBE8 W5KQY5 A0A1A9UP56 A0A0A1WYP5 A0A3B4B8T0 Q17ML5 A0A151X3K9 A0A1W2WB15 A0A151I2C1 A0A158NKZ4 A0A1B8Y939 Q6NX31 A0A0A9XGX9 F4WWT5 A0A1B0CLF2 E2BME7 A0A1Y1KVC9 A0A0J7KE46 A0A3B3RCA0 A0A1B0FC91 A0A0P4VTB0 A0A3Q0R3X1 A0A0C9QWH7 A0A1A9ZBZ2 A0A146XPY6 A0A423TWM4 A0A182GAT2 A0A1A9XQV9 A0A1B0ANR5 A0A1A9W3H0 B0VZP1 A0A084WDI5 Q91876 A0A087UTA1 A0A3B5KJS6 A0A2D0PT24 A0A3Q1BNY5 A0A3B4Z553 A0A2L2Y8F0
A0A0L7KNN6 A0A194RP00 D6WK36 A0A2J7QTN3 A0A1B6CHG2 A0A090XB26 B4HK41 A0A131XVG5 A0A067R721 A0A131Z543 A0A1E1XG68 A0A2P8Y7I5 L7M6G7 A0A224YVW3 A0A1B6MDL9 A0A336MAI7 B4H9M2 A0A293N4B1 A0A1Z5KUK0 A0A1B6IDU9 B4QML9 B3NBM4 B4PFH7 Q9XYU0 B4KZG3 B7QHT6 A0A1W4VCI4 A0A0J9RS32 A0A3R7QWK7 A0A1B6GR93 Q29D32 E2AL70 A0A090XAY7 C3ZEX3 A0A3Q0ITS1 A0A3B0JBE8 B4LEP6 A0A1I8Q0B2 A0A0L0BMS2 A0A1J1IEB8 A0A0M4EGR2 A0A026WPZ1 N6TF82 W8BNL9 V5KXP1 B3M5X9 A0A0K8WEX4 A0A2P2I1N8 A0A151K3E9 A0A1L8H3C0 Q7ZXB1 B4MMD1 B4J3L8 A0A195FTX8 T1PBE8 W5KQY5 A0A1A9UP56 A0A0A1WYP5 A0A3B4B8T0 Q17ML5 A0A151X3K9 A0A1W2WB15 A0A151I2C1 A0A158NKZ4 A0A1B8Y939 Q6NX31 A0A0A9XGX9 F4WWT5 A0A1B0CLF2 E2BME7 A0A1Y1KVC9 A0A0J7KE46 A0A3B3RCA0 A0A1B0FC91 A0A0P4VTB0 A0A3Q0R3X1 A0A0C9QWH7 A0A1A9ZBZ2 A0A146XPY6 A0A423TWM4 A0A182GAT2 A0A1A9XQV9 A0A1B0ANR5 A0A1A9W3H0 B0VZP1 A0A084WDI5 Q91876 A0A087UTA1 A0A3B5KJS6 A0A2D0PT24 A0A3Q1BNY5 A0A3B4Z553 A0A2L2Y8F0
PDB
6HV9
E-value=4.38903e-10,
Score=150
Ontologies
GO
PANTHER
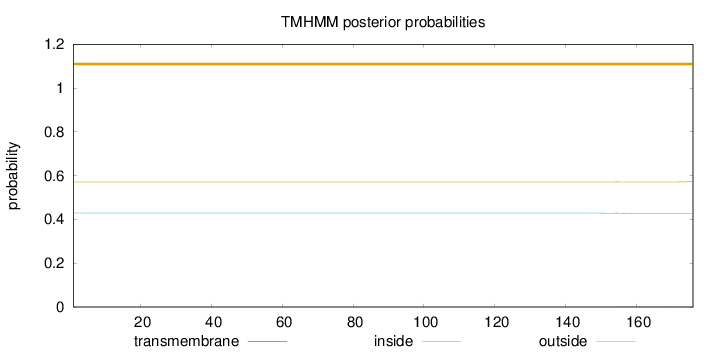
Topology
Subcellular location
Nucleus
Length:
176
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03568
Exp number, first 60 AAs:
0
Total prob of N-in:
0.42874
outside
1 - 176
Population Genetic Test Statistics
Pi
247.768273
Theta
202.194401
Tajima's D
0.831629
CLR
0
CSRT
0.615269236538173
Interpretation
Uncertain
