Gene
KWMTBOMO09197
Pre Gene Modal
BGIBMGA003353
Annotation
PREDICTED:_glypican-5_[Papilio_xuthus]
Full name
Division abnormally delayed protein
Location in the cell
Nuclear Reliability : 2.225
Sequence
CDS
ATGCTCTCCACCAGGCGGACGTTGCAAGAGCATCTCTTGGGACTATCGCATCGATCTCAAAACAAAACAGCGGTACTCTTCACGCAATTGTACCGCGGGCACGCGACGAGGACTCACACTCCTCTGGCGACCCTCTATGACGACATACGAACTCTCCTACGATCGAGTTCCGAGCATGACGTCATAACCGACATCGTCAACACGAGGCATCCTAAAGATTTGGTCACCAGCGCAAGGAAGTTCTTCAGAGATCTATTCCCGGTTGCGTATCAAAACGCACTCAGACTAGACTCGAAGCAGTTCACGCCGGAGTACGAAGCCTGCCTCAAAGATGCCTATGACGCCGTCCAGCCCTTCGGAGAAGTTCCGCAACAGCTCGGCGTTTCCCTGTCTCAGTCCCTGGAGGCAGCGCGAGTGCTCTTGGACATGCTCGCGGTGGGCGCCGGCGCTCTGACCTCCGCCAACCACGTGCTCGCCGGTATTAACGAGGAATGCTCCAACAAGCTACTCCAGGCCGCTGGGTGTGCGCGCTGCGCTGGATTCGACGCGAGACCATGCAGGAACTACTGCTTGAATATCGCTCGAGGGTGTATTGGATCTCTCTTGGTAGAGCTAGACGGGCCATGGTCTAGCTACGTGGAAGGAGTGGAGCAGCTCACCAGGATAGATGCTGACGTCGCACTACGAGAACTCGACACCAAAGTATCAAAAGCAATAATGTACGCTCTCGAGAACAGAAACATTTTGGAGAATAAGGTACGTCAAGAATGTGGTCCTCCGTCGACTGTGGACTCCGGTCTCACGCCGGCCCCACCCACGCCTGGTTCGGTGAGACGAGACGACTTCCGAGCGCCGCCTCCGGACACGGAGCTTCTCCAATTCGCGGCGACCCTCGCTTCCACAAAGAGGCTGTTCAGCACTGTCGCGGACAGGCTGTGTGACGAGACCGACTTCGTTGATGACGTCAATGAACACTGTTGGAACGGACAGGCCATTGGCGAATACTCGAAACCATTGGTGCCTTCATCGACACTTAGCAACCAGAAGTATAATCCTGAGATCACACTGAACCCACCACAGGACCCGCGTCTTCTTAACATTGCCGATAGACTGCAGCAAGCCAGACAGTTGCTGATCTCACACGCGGGGACCGGGGGGGCGCAAGCTGAGGCTTTCATGCAGGGAGATGAGGCAGGCGAAGAAGGGTCCGGCTCTGGTCGAAGTTATCCCGAAGACGACGGATCTTATGACGCCGAGGGTTCAGGTGAAGAGGGATCTGGCTCCGACGACGATGTGAGCAGAACAATGGCCGGCGAGAATCGAGGAGGCTCCGCTACGTATTCTTCGACTGAACGAACGCAAGCCGCCACTAACACACGACCATTCCTCTCAGTTATATTTGCAACACTGATCTTAGCTGCTTTTGGACAATGCGTTACATAA
Protein
MLSTRRTLQEHLLGLSHRSQNKTAVLFTQLYRGHATRTHTPLATLYDDIRTLLRSSSEHDVITDIVNTRHPKDLVTSARKFFRDLFPVAYQNALRLDSKQFTPEYEACLKDAYDAVQPFGEVPQQLGVSLSQSLEAARVLLDMLAVGAGALTSANHVLAGINEECSNKLLQAAGCARCAGFDARPCRNYCLNIARGCIGSLLVELDGPWSSYVEGVEQLTRIDADVALRELDTKVSKAIMYALENRNILENKVRQECGPPSTVDSGLTPAPPTPGSVRRDDFRAPPPDTELLQFAATLASTKRLFSTVADRLCDETDFVDDVNEHCWNGQAIGEYSKPLVPSSTLSNQKYNPEITLNPPQDPRLLNIADRLQQARQLLISHAGTGGAQAEAFMQGDEAGEEGSGSGRSYPEDDGSYDAEGSGEEGSGSDDDVSRTMAGENRGGSATYSSTERTQAATNTRPFLSVIFATLILAAFGQCVT
Summary
Description
Cell surface proteoglycan that bears heparan sulfate.
Similarity
Belongs to the glypican family.
Keywords
Cell membrane
Complete proteome
Glycoprotein
GPI-anchor
Heparan sulfate
Lipoprotein
Membrane
Proteoglycan
Reference proteome
Signal
Feature
chain Division abnormally delayed protein
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
A0A2A4K4B7
A0A212EZZ9
A0A194PLC3
A0A3S2NAZ7
A0A0N1IAM2
A0A1Y1NHI2
+ More
A0A139WHC0 V5GRD8 A0A0L0BMF3 U5ESP5 A0A1A9UP57 A0A1B0FBP7 W8C359 A0A0K8UY79 A0A0A1XKY6 A0A087UDT6 A0A151I248 A0A1I8PGG7 A0A151X3L3 A0A1L8DJF1 A0A158NKZ6 A0A1L8DK29 B4KZG2 B4LEP5 A0A1A9W3G9 A0A182HB33 A0A0C9PZC5 A0A1A9ZBZ3 A0A1A9XQV8 A0A1B0ANR8 A0A1B6LF05 A0A1B6DS00 A0A3L8DJU7 B4J3L9 E0VBY5 A0A336M3A7 T1P7N7 A0A1I8NB62 A0A1I8N9J2 A0A3B0J5R3 A0A2K8JS74 K7IXM2 A0A195CB27 A0A1W4VCM7 A0A195FSC9 Q29D31 A0A1S3HQQ8 A0A1S3JSX1 B3M5Y0 Q17ML3 A0A084WDI6 A0A1J1HX35 A0A224XM98 B4MMD2 A0A1Q3G519 A0A1Q3G509 A0A1Q3G504 A0A151J2U5 A0A1Q3G507 A0A1Q3G513 A0A1Q3G508 A0A0J9RT49 A0A1Q3G4Z4 A0A1Q3G514 Q966V5 Q53XG2 Q24114 B4HK40 A0A1Q3G510 A0A2S2NHB4 A0A0M3QWW2 A0A026WMB4 A0A336M7C5 A0A443ST61 B3NBM5 J9JYH8 A0A2M4BHB6 A0A2M4BGP7 B4PFH8 A0A2H8TKV7 A0A2M4DKZ8 B4QML8 A0A154PF46 A0A0N8D4A1 A0A2C9H5S9 A0A0P5U8G9 A0A0N8DFE9 A0A0P6IQ30 Q7Q6K2 A0A348G6D0
A0A139WHC0 V5GRD8 A0A0L0BMF3 U5ESP5 A0A1A9UP57 A0A1B0FBP7 W8C359 A0A0K8UY79 A0A0A1XKY6 A0A087UDT6 A0A151I248 A0A1I8PGG7 A0A151X3L3 A0A1L8DJF1 A0A158NKZ6 A0A1L8DK29 B4KZG2 B4LEP5 A0A1A9W3G9 A0A182HB33 A0A0C9PZC5 A0A1A9ZBZ3 A0A1A9XQV8 A0A1B0ANR8 A0A1B6LF05 A0A1B6DS00 A0A3L8DJU7 B4J3L9 E0VBY5 A0A336M3A7 T1P7N7 A0A1I8NB62 A0A1I8N9J2 A0A3B0J5R3 A0A2K8JS74 K7IXM2 A0A195CB27 A0A1W4VCM7 A0A195FSC9 Q29D31 A0A1S3HQQ8 A0A1S3JSX1 B3M5Y0 Q17ML3 A0A084WDI6 A0A1J1HX35 A0A224XM98 B4MMD2 A0A1Q3G519 A0A1Q3G509 A0A1Q3G504 A0A151J2U5 A0A1Q3G507 A0A1Q3G513 A0A1Q3G508 A0A0J9RT49 A0A1Q3G4Z4 A0A1Q3G514 Q966V5 Q53XG2 Q24114 B4HK40 A0A1Q3G510 A0A2S2NHB4 A0A0M3QWW2 A0A026WMB4 A0A336M7C5 A0A443ST61 B3NBM5 J9JYH8 A0A2M4BHB6 A0A2M4BGP7 B4PFH8 A0A2H8TKV7 A0A2M4DKZ8 B4QML8 A0A154PF46 A0A0N8D4A1 A0A2C9H5S9 A0A0P5U8G9 A0A0N8DFE9 A0A0P6IQ30 Q7Q6K2 A0A348G6D0
Pubmed
22118469
26354079
28004739
18362917
19820115
26108605
+ More
24495485 25830018 21347285 17994087 26483478 30249741 20566863 25315136 20075255 15632085 26383154 17510324 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8582281 19349973 24508170 17550304 12364791 14747013 17210077
24495485 25830018 21347285 17994087 26483478 30249741 20566863 25315136 20075255 15632085 26383154 17510324 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8582281 19349973 24508170 17550304 12364791 14747013 17210077
EMBL
NWSH01000144
PCG79105.1
AGBW02011215
OWR47037.1
KQ459602
KPI93534.1
+ More
RSAL01000296 RVE42820.1 KQ460317 KPJ15986.1 GEZM01006895 JAV95586.1 KQ971343 KYB27316.1 GALX01004294 JAB64172.1 JRES01001641 KNC21310.1 GANO01003147 JAB56724.1 CCAG010004103 GAMC01010306 GAMC01010301 JAB96254.1 GDHF01020816 JAI31498.1 GBXI01003089 JAD11203.1 KK119378 KFM75525.1 KQ976546 KYM80960.1 KQ982557 KYQ55027.1 GFDF01007587 JAV06497.1 ADTU01019058 ADTU01019059 ADTU01019060 ADTU01019061 ADTU01019062 ADTU01019063 ADTU01019064 GFDF01007313 JAV06771.1 CH933809 EDW17889.1 CH940647 EDW69130.2 JXUM01006264 KQ560212 KXJ83819.1 GBYB01006882 GBYB01006883 JAG76649.1 JAG76650.1 JXJN01000933 GEBQ01017724 JAT22253.1 GEDC01008847 JAS28451.1 QOIP01000007 RLU20690.1 CH916366 EDV96221.1 DS235044 EEB10891.1 UFQS01000494 UFQT01000494 SSX04368.1 SSX24732.1 KA644512 AFP59141.1 OUUW01000002 SPP77354.1 KY031139 ATU82890.1 AAZX01000021 AAZX01000038 AAZX01011155 KQ978072 KYM97313.1 KQ981281 KYN43346.1 CH379070 EAL30583.3 CH902618 EDV39670.1 CH477205 EAT47911.1 ATLV01023023 ATLV01023024 ATLV01023025 KE525339 KFB48280.1 CVRI01000035 CRK92663.1 GFTR01007285 JAW09141.1 CH963847 EDW73277.2 GFDL01000151 JAV34894.1 GFDL01000161 JAV34884.1 GFDL01000153 JAV34892.1 KQ980342 KYN16370.1 GFDL01000157 JAV34888.1 GFDL01000158 JAV34887.1 GFDL01000160 JAV34885.1 CM002912 KMY98489.1 GFDL01000163 JAV34882.1 GFDL01000162 JAV34883.1 AB052367 BAB60703.1 BT011348 AE014296 AAR96140.1 AFH04356.1 U31985 CH480815 EDW40776.1 GFDL01000155 JAV34890.1 GGMR01003972 MBY16591.1 CP012525 ALC44873.1 KK107152 EZA57155.1 SSX04369.1 SSX24733.1 NCKV01000403 RWS30717.1 CH954178 EDV50621.1 ABLF02031508 ABLF02031517 ABLF02031519 ABLF02031525 ABLF02031526 GGFJ01003242 MBW52383.1 GGFJ01002980 MBW52121.1 CM000159 EDW93104.1 GFXV01002970 MBW14775.1 GGFL01014046 MBW78224.1 CM000363 EDX09772.1 KQ434885 KZC10064.1 GDIP01070239 JAM33476.1 GDIP01116576 JAL87138.1 GDIP01039055 JAM64660.1 GDIQ01028656 JAN66081.1 AAAB01008960 EAA10793.4 EGK97281.1 FX986046 BBF98003.1
RSAL01000296 RVE42820.1 KQ460317 KPJ15986.1 GEZM01006895 JAV95586.1 KQ971343 KYB27316.1 GALX01004294 JAB64172.1 JRES01001641 KNC21310.1 GANO01003147 JAB56724.1 CCAG010004103 GAMC01010306 GAMC01010301 JAB96254.1 GDHF01020816 JAI31498.1 GBXI01003089 JAD11203.1 KK119378 KFM75525.1 KQ976546 KYM80960.1 KQ982557 KYQ55027.1 GFDF01007587 JAV06497.1 ADTU01019058 ADTU01019059 ADTU01019060 ADTU01019061 ADTU01019062 ADTU01019063 ADTU01019064 GFDF01007313 JAV06771.1 CH933809 EDW17889.1 CH940647 EDW69130.2 JXUM01006264 KQ560212 KXJ83819.1 GBYB01006882 GBYB01006883 JAG76649.1 JAG76650.1 JXJN01000933 GEBQ01017724 JAT22253.1 GEDC01008847 JAS28451.1 QOIP01000007 RLU20690.1 CH916366 EDV96221.1 DS235044 EEB10891.1 UFQS01000494 UFQT01000494 SSX04368.1 SSX24732.1 KA644512 AFP59141.1 OUUW01000002 SPP77354.1 KY031139 ATU82890.1 AAZX01000021 AAZX01000038 AAZX01011155 KQ978072 KYM97313.1 KQ981281 KYN43346.1 CH379070 EAL30583.3 CH902618 EDV39670.1 CH477205 EAT47911.1 ATLV01023023 ATLV01023024 ATLV01023025 KE525339 KFB48280.1 CVRI01000035 CRK92663.1 GFTR01007285 JAW09141.1 CH963847 EDW73277.2 GFDL01000151 JAV34894.1 GFDL01000161 JAV34884.1 GFDL01000153 JAV34892.1 KQ980342 KYN16370.1 GFDL01000157 JAV34888.1 GFDL01000158 JAV34887.1 GFDL01000160 JAV34885.1 CM002912 KMY98489.1 GFDL01000163 JAV34882.1 GFDL01000162 JAV34883.1 AB052367 BAB60703.1 BT011348 AE014296 AAR96140.1 AFH04356.1 U31985 CH480815 EDW40776.1 GFDL01000155 JAV34890.1 GGMR01003972 MBY16591.1 CP012525 ALC44873.1 KK107152 EZA57155.1 SSX04369.1 SSX24733.1 NCKV01000403 RWS30717.1 CH954178 EDV50621.1 ABLF02031508 ABLF02031517 ABLF02031519 ABLF02031525 ABLF02031526 GGFJ01003242 MBW52383.1 GGFJ01002980 MBW52121.1 CM000159 EDW93104.1 GFXV01002970 MBW14775.1 GGFL01014046 MBW78224.1 CM000363 EDX09772.1 KQ434885 KZC10064.1 GDIP01070239 JAM33476.1 GDIP01116576 JAL87138.1 GDIP01039055 JAM64660.1 GDIQ01028656 JAN66081.1 AAAB01008960 EAA10793.4 EGK97281.1 FX986046 BBF98003.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000283053
UP000053240
UP000007266
+ More
UP000037069 UP000078200 UP000092444 UP000054359 UP000078540 UP000095300 UP000075809 UP000005205 UP000009192 UP000008792 UP000091820 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000279307 UP000001070 UP000009046 UP000095301 UP000268350 UP000002358 UP000078542 UP000192221 UP000078541 UP000001819 UP000085678 UP000007801 UP000008820 UP000030765 UP000183832 UP000007798 UP000078492 UP000000803 UP000001292 UP000092553 UP000053097 UP000288716 UP000008711 UP000007819 UP000002282 UP000000304 UP000076502 UP000075903 UP000007062
UP000037069 UP000078200 UP000092444 UP000054359 UP000078540 UP000095300 UP000075809 UP000005205 UP000009192 UP000008792 UP000091820 UP000069940 UP000249989 UP000092445 UP000092443 UP000092460 UP000279307 UP000001070 UP000009046 UP000095301 UP000268350 UP000002358 UP000078542 UP000192221 UP000078541 UP000001819 UP000085678 UP000007801 UP000008820 UP000030765 UP000183832 UP000007798 UP000078492 UP000000803 UP000001292 UP000092553 UP000053097 UP000288716 UP000008711 UP000007819 UP000002282 UP000000304 UP000076502 UP000075903 UP000007062
Pfam
PF01153 Glypican
Gene 3D
ProteinModelPortal
A0A2A4K4B7
A0A212EZZ9
A0A194PLC3
A0A3S2NAZ7
A0A0N1IAM2
A0A1Y1NHI2
+ More
A0A139WHC0 V5GRD8 A0A0L0BMF3 U5ESP5 A0A1A9UP57 A0A1B0FBP7 W8C359 A0A0K8UY79 A0A0A1XKY6 A0A087UDT6 A0A151I248 A0A1I8PGG7 A0A151X3L3 A0A1L8DJF1 A0A158NKZ6 A0A1L8DK29 B4KZG2 B4LEP5 A0A1A9W3G9 A0A182HB33 A0A0C9PZC5 A0A1A9ZBZ3 A0A1A9XQV8 A0A1B0ANR8 A0A1B6LF05 A0A1B6DS00 A0A3L8DJU7 B4J3L9 E0VBY5 A0A336M3A7 T1P7N7 A0A1I8NB62 A0A1I8N9J2 A0A3B0J5R3 A0A2K8JS74 K7IXM2 A0A195CB27 A0A1W4VCM7 A0A195FSC9 Q29D31 A0A1S3HQQ8 A0A1S3JSX1 B3M5Y0 Q17ML3 A0A084WDI6 A0A1J1HX35 A0A224XM98 B4MMD2 A0A1Q3G519 A0A1Q3G509 A0A1Q3G504 A0A151J2U5 A0A1Q3G507 A0A1Q3G513 A0A1Q3G508 A0A0J9RT49 A0A1Q3G4Z4 A0A1Q3G514 Q966V5 Q53XG2 Q24114 B4HK40 A0A1Q3G510 A0A2S2NHB4 A0A0M3QWW2 A0A026WMB4 A0A336M7C5 A0A443ST61 B3NBM5 J9JYH8 A0A2M4BHB6 A0A2M4BGP7 B4PFH8 A0A2H8TKV7 A0A2M4DKZ8 B4QML8 A0A154PF46 A0A0N8D4A1 A0A2C9H5S9 A0A0P5U8G9 A0A0N8DFE9 A0A0P6IQ30 Q7Q6K2 A0A348G6D0
A0A139WHC0 V5GRD8 A0A0L0BMF3 U5ESP5 A0A1A9UP57 A0A1B0FBP7 W8C359 A0A0K8UY79 A0A0A1XKY6 A0A087UDT6 A0A151I248 A0A1I8PGG7 A0A151X3L3 A0A1L8DJF1 A0A158NKZ6 A0A1L8DK29 B4KZG2 B4LEP5 A0A1A9W3G9 A0A182HB33 A0A0C9PZC5 A0A1A9ZBZ3 A0A1A9XQV8 A0A1B0ANR8 A0A1B6LF05 A0A1B6DS00 A0A3L8DJU7 B4J3L9 E0VBY5 A0A336M3A7 T1P7N7 A0A1I8NB62 A0A1I8N9J2 A0A3B0J5R3 A0A2K8JS74 K7IXM2 A0A195CB27 A0A1W4VCM7 A0A195FSC9 Q29D31 A0A1S3HQQ8 A0A1S3JSX1 B3M5Y0 Q17ML3 A0A084WDI6 A0A1J1HX35 A0A224XM98 B4MMD2 A0A1Q3G519 A0A1Q3G509 A0A1Q3G504 A0A151J2U5 A0A1Q3G507 A0A1Q3G513 A0A1Q3G508 A0A0J9RT49 A0A1Q3G4Z4 A0A1Q3G514 Q966V5 Q53XG2 Q24114 B4HK40 A0A1Q3G510 A0A2S2NHB4 A0A0M3QWW2 A0A026WMB4 A0A336M7C5 A0A443ST61 B3NBM5 J9JYH8 A0A2M4BHB6 A0A2M4BGP7 B4PFH8 A0A2H8TKV7 A0A2M4DKZ8 B4QML8 A0A154PF46 A0A0N8D4A1 A0A2C9H5S9 A0A0P5U8G9 A0A0N8DFE9 A0A0P6IQ30 Q7Q6K2 A0A348G6D0
PDB
4BWE
E-value=1.26674e-23,
Score=273
Ontologies
PATHWAY
GO
GO:0062023
GO:0008101
GO:0009966
GO:0016055
GO:0046658
GO:0031225
GO:0005886
GO:0016021
GO:0005576
GO:0009986
GO:0090263
GO:0016477
GO:1905475
GO:0045743
GO:0071694
GO:0030718
GO:0030513
GO:0048190
GO:0008407
GO:0007367
GO:0007480
GO:0048132
GO:2001261
GO:0040014
GO:0007427
GO:0008045
GO:0031362
GO:0016324
GO:0045880
GO:0008052
GO:0008586
GO:0048813
GO:0051726
GO:0045570
GO:0016323
GO:0048749
GO:0007224
GO:0007409
GO:0035222
GO:0030510
GO:0007472
GO:0035220
GO:0022416
GO:0098595
GO:0005578
GO:0016020
GO:0043395
GO:0005524
GO:0005739
GO:0016776
GO:0006811
GO:0016876
GO:0043039
PANTHER
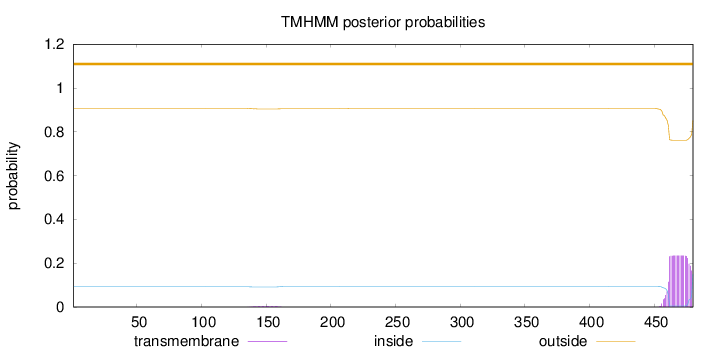
Topology
Subcellular location
Cell membrane
Length:
480
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.47539
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.09235
outside
1 - 480
Population Genetic Test Statistics
Pi
235.219002
Theta
177.186682
Tajima's D
1.189079
CLR
0.13747
CSRT
0.714114294285286
Interpretation
Uncertain
