Gene
KWMTBOMO09190 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003358
Annotation
PREDICTED:_HIG1_domain_family_member_1A?_mitochondrial-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.342 Nuclear Reliability : 1.004
Sequence
CDS
ATGACTAACAGCCAACAAATATTTGAATACAATGAAGAATCACAATCTGAAAGACTTGCCAGGAAGTCAAAAGATTCACCATTTATGATAATTGGGCTCCTTGGATTAGTAGGTGTTTGTGGCTATGGTGCATACAGTTACAAGAAGAGAGGAAAAATGAGCACTAGTGTATTCCTAATGCAGCTCCGAGTTGCTGCCCAAGGCACTGTAGTGTCAGCTCTTACAATCGGGCTGGCATACACATTAGCAAAGGAACACCTCTTTAAAGATTCTAAGAAAGAATAA
Protein
MTNSQQIFEYNEESQSERLARKSKDSPFMIIGLLGLVGVCGYGAYSYKKRGKMSTSVFLMQLRVAAQGTVVSALTIGLAYTLAKEHLFKDSKKE
Summary
Uniprot
H9J1H1
A0A3S2N6M0
A0A2H1V2K9
A0A2A4J6H5
I4DK40
A0A194R7I3
+ More
S4NSW2 A0A1L8E0J0 H9J1G9 A0A1B0DQY2 A0A0L7KNI9 A0A1B0CJ30 A0A0K8TRA6 B4NKV9 A0A084WNK2 U5EHM4 A0A023ECJ9 A0A336L9C0 B0XDC9 A0A2H1V2D6 Q16XY6 T1E2F2 Q16XY7 W8BMG1 A0A1Y1MKT1 A0A023EFU6 A0A182N2N3 A0A194RD06 A0A194PJD0 A0A1Q3FIB5 A0A182TEC5 A0A182WXW7 Q7QE07 A0A182I5Y8 A0A182FVG1 A0A2M4AGW9 T1DQL4 W5JI78 A0A182WE50 A0A182Y9M2 A0A2M4C584 A0A2M4C429 A0A182VHQ2 A0A182ISZ0 Q293S5 B4GLP6 A0A336MUQ4 A0A1W4V5A7 A0A182PM22 A0A182K180 A0A3B0JNX6 A0A0A1WG42 R4UKW7 A0A2M3ZKK1 Q9VGQ2 A0A0K8UPN9 A0A034WJ12 A0A0C9QUI6 Q8INK7 B4QID4 B3NZG3 A0A2M4B0D7 B4HMP5 A0A2A4J5S6 A0A0P9AQR0 B3LXD6 Q8SXE6 A0A1E1WDW2 A0A0Q9XA11 B4K9U8 R4WNN7 B5RIK2 A0A0Q9XD25 B4QU55 A0A1W4WUF7 A0A1W4WU50 A0A1W4WT46 A0A0J9R9N6 B4PM41 D6WIG9 A0A1E1WM56 A0A067QPA3 N6TG59 A0A212F015 Q7JW46 B4HIB1 A0A0M4EEG2 A0A0Q9WFS3 B4LXM6 A0A1J1J4G3 T1I5P0 A0A0B4KEI1 A0A1W4VQ53 A0A023FCS0 A0A0V0GBZ0 A0A0R1DMZ5 A0A1B0G4T7 A0A2P8Z1R7 A0A240SX62 A0A1B0AZE3
S4NSW2 A0A1L8E0J0 H9J1G9 A0A1B0DQY2 A0A0L7KNI9 A0A1B0CJ30 A0A0K8TRA6 B4NKV9 A0A084WNK2 U5EHM4 A0A023ECJ9 A0A336L9C0 B0XDC9 A0A2H1V2D6 Q16XY6 T1E2F2 Q16XY7 W8BMG1 A0A1Y1MKT1 A0A023EFU6 A0A182N2N3 A0A194RD06 A0A194PJD0 A0A1Q3FIB5 A0A182TEC5 A0A182WXW7 Q7QE07 A0A182I5Y8 A0A182FVG1 A0A2M4AGW9 T1DQL4 W5JI78 A0A182WE50 A0A182Y9M2 A0A2M4C584 A0A2M4C429 A0A182VHQ2 A0A182ISZ0 Q293S5 B4GLP6 A0A336MUQ4 A0A1W4V5A7 A0A182PM22 A0A182K180 A0A3B0JNX6 A0A0A1WG42 R4UKW7 A0A2M3ZKK1 Q9VGQ2 A0A0K8UPN9 A0A034WJ12 A0A0C9QUI6 Q8INK7 B4QID4 B3NZG3 A0A2M4B0D7 B4HMP5 A0A2A4J5S6 A0A0P9AQR0 B3LXD6 Q8SXE6 A0A1E1WDW2 A0A0Q9XA11 B4K9U8 R4WNN7 B5RIK2 A0A0Q9XD25 B4QU55 A0A1W4WUF7 A0A1W4WU50 A0A1W4WT46 A0A0J9R9N6 B4PM41 D6WIG9 A0A1E1WM56 A0A067QPA3 N6TG59 A0A212F015 Q7JW46 B4HIB1 A0A0M4EEG2 A0A0Q9WFS3 B4LXM6 A0A1J1J4G3 T1I5P0 A0A0B4KEI1 A0A1W4VQ53 A0A023FCS0 A0A0V0GBZ0 A0A0R1DMZ5 A0A1B0G4T7 A0A2P8Z1R7 A0A240SX62 A0A1B0AZE3
Pubmed
19121390
22651552
26354079
23622113
26227816
26369729
+ More
17994087 24438588 24945155 26483478 17510324 24330624 24495485 28004739 12364791 14747013 17210077 20920257 23761445 25244985 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 23691247 22936249 17550304 18362917 19820115 24845553 23537049 22118469 25474469 29403074
17994087 24438588 24945155 26483478 17510324 24330624 24495485 28004739 12364791 14747013 17210077 20920257 23761445 25244985 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 23691247 22936249 17550304 18362917 19820115 24845553 23537049 22118469 25474469 29403074
EMBL
BABH01023748
RSAL01000296
RVE42815.1
ODYU01000358
SOQ35006.1
NWSH01003059
+ More
PCG67023.1 AK401658 KQ459602 BAM18280.1 KPI93528.1 KQ460615 KPJ13788.1 GAIX01013882 JAA78678.1 GFDF01002069 JAV12015.1 AJVK01019452 JTDY01007915 KOB64857.1 AJWK01013987 AJWK01013988 GDAI01001148 JAI16455.1 CH964272 EDW84162.1 ATLV01024619 KE525353 KFB51796.1 GANO01002954 JAB56917.1 JXUM01034371 GAPW01006530 KQ561021 JAC07068.1 KXJ79999.1 UFQS01002436 UFQT01002436 SSX14029.1 SSX33448.1 DS232752 EDS45407.1 SOQ35003.1 CH477530 EAT39456.1 GALA01001300 JAA93552.1 EAT39457.1 GAMC01008388 GAMC01008387 JAB98168.1 GEZM01031222 JAV84965.1 GAPW01005898 JAC07700.1 KPJ13791.1 KPI93531.1 GFDL01007802 JAV27243.1 AAAB01008848 EAA07070.3 APCN01003537 GGFK01006641 MBW39962.1 GAMD01001095 JAB00496.1 ADMH02001177 ETN63831.1 GGFJ01011170 MBW60311.1 GGFJ01010903 MBW60044.1 CM000070 EAL29139.2 CH479185 EDW38470.1 SSX14026.1 SSX33445.1 OUUW01000008 SPP83825.1 GBXI01016475 JAC97816.1 KC740955 AGM32779.1 GGFM01008310 MBW29061.1 AE014297 AY070609 AAF54624.1 AAL48080.1 GDHF01023667 JAI28647.1 GAKP01004625 JAC54327.1 GBYB01004297 JAG74064.1 KX531469 AAN13498.1 ANY27279.1 CM000362 EDX06494.1 CH954181 EDV49536.1 GGFK01013169 MBW46490.1 CH480816 EDW47257.1 PCG67028.1 CH902617 KPU79902.1 EDV42780.1 AY089682 AAL90420.1 GDQN01006003 JAT85051.1 CH933806 KRG01634.1 EDW15596.1 AK417271 BAN20486.1 BT044126 ACH92191.1 KRG01635.1 CM000364 EDX13386.1 CM002911 KMY92783.1 CM000160 EDW96907.1 KQ971334 EFA01076.1 GDQN01003047 JAT88007.1 KK853110 KDR11232.1 APGK01002433 APGK01004119 APGK01039325 KB740969 KB735583 KB734777 KB632230 ENN76753.1 ENN83343.1 ENN83504.1 ERL90393.1 AGBW02011215 OWR47034.1 AE013599 AY118378 AAF58798.1 AAM48407.1 CH480815 EDW42626.1 CP012526 ALC46378.1 CH940650 KRF82934.1 EDW66810.1 CVRI01000067 CRL06692.1 ACPB03003574 AGB93398.1 GBBI01000263 JAC18449.1 GECL01000767 JAP05357.1 CM000157 KRJ98644.1 KRJ98645.1 CCAG010006284 PYGN01000238 PSN50447.1 JXJN01006255
PCG67023.1 AK401658 KQ459602 BAM18280.1 KPI93528.1 KQ460615 KPJ13788.1 GAIX01013882 JAA78678.1 GFDF01002069 JAV12015.1 AJVK01019452 JTDY01007915 KOB64857.1 AJWK01013987 AJWK01013988 GDAI01001148 JAI16455.1 CH964272 EDW84162.1 ATLV01024619 KE525353 KFB51796.1 GANO01002954 JAB56917.1 JXUM01034371 GAPW01006530 KQ561021 JAC07068.1 KXJ79999.1 UFQS01002436 UFQT01002436 SSX14029.1 SSX33448.1 DS232752 EDS45407.1 SOQ35003.1 CH477530 EAT39456.1 GALA01001300 JAA93552.1 EAT39457.1 GAMC01008388 GAMC01008387 JAB98168.1 GEZM01031222 JAV84965.1 GAPW01005898 JAC07700.1 KPJ13791.1 KPI93531.1 GFDL01007802 JAV27243.1 AAAB01008848 EAA07070.3 APCN01003537 GGFK01006641 MBW39962.1 GAMD01001095 JAB00496.1 ADMH02001177 ETN63831.1 GGFJ01011170 MBW60311.1 GGFJ01010903 MBW60044.1 CM000070 EAL29139.2 CH479185 EDW38470.1 SSX14026.1 SSX33445.1 OUUW01000008 SPP83825.1 GBXI01016475 JAC97816.1 KC740955 AGM32779.1 GGFM01008310 MBW29061.1 AE014297 AY070609 AAF54624.1 AAL48080.1 GDHF01023667 JAI28647.1 GAKP01004625 JAC54327.1 GBYB01004297 JAG74064.1 KX531469 AAN13498.1 ANY27279.1 CM000362 EDX06494.1 CH954181 EDV49536.1 GGFK01013169 MBW46490.1 CH480816 EDW47257.1 PCG67028.1 CH902617 KPU79902.1 EDV42780.1 AY089682 AAL90420.1 GDQN01006003 JAT85051.1 CH933806 KRG01634.1 EDW15596.1 AK417271 BAN20486.1 BT044126 ACH92191.1 KRG01635.1 CM000364 EDX13386.1 CM002911 KMY92783.1 CM000160 EDW96907.1 KQ971334 EFA01076.1 GDQN01003047 JAT88007.1 KK853110 KDR11232.1 APGK01002433 APGK01004119 APGK01039325 KB740969 KB735583 KB734777 KB632230 ENN76753.1 ENN83343.1 ENN83504.1 ERL90393.1 AGBW02011215 OWR47034.1 AE013599 AY118378 AAF58798.1 AAM48407.1 CH480815 EDW42626.1 CP012526 ALC46378.1 CH940650 KRF82934.1 EDW66810.1 CVRI01000067 CRL06692.1 ACPB03003574 AGB93398.1 GBBI01000263 JAC18449.1 GECL01000767 JAP05357.1 CM000157 KRJ98644.1 KRJ98645.1 CCAG010006284 PYGN01000238 PSN50447.1 JXJN01006255
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000092462
+ More
UP000037510 UP000092461 UP000007798 UP000030765 UP000069940 UP000249989 UP000002320 UP000008820 UP000075884 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000000673 UP000075920 UP000076408 UP000075903 UP000075880 UP000001819 UP000008744 UP000192221 UP000075885 UP000075881 UP000268350 UP000000803 UP000000304 UP000008711 UP000001292 UP000007801 UP000009192 UP000192223 UP000002282 UP000007266 UP000027135 UP000019118 UP000030742 UP000007151 UP000092553 UP000008792 UP000183832 UP000015103 UP000092444 UP000245037 UP000092445 UP000092460
UP000037510 UP000092461 UP000007798 UP000030765 UP000069940 UP000249989 UP000002320 UP000008820 UP000075884 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000000673 UP000075920 UP000076408 UP000075903 UP000075880 UP000001819 UP000008744 UP000192221 UP000075885 UP000075881 UP000268350 UP000000803 UP000000304 UP000008711 UP000001292 UP000007801 UP000009192 UP000192223 UP000002282 UP000007266 UP000027135 UP000019118 UP000030742 UP000007151 UP000092553 UP000008792 UP000183832 UP000015103 UP000092444 UP000245037 UP000092445 UP000092460
Pfam
PF04588 HIG_1_N
Interpro
IPR007667
Hypoxia_induced_domain
ProteinModelPortal
H9J1H1
A0A3S2N6M0
A0A2H1V2K9
A0A2A4J6H5
I4DK40
A0A194R7I3
+ More
S4NSW2 A0A1L8E0J0 H9J1G9 A0A1B0DQY2 A0A0L7KNI9 A0A1B0CJ30 A0A0K8TRA6 B4NKV9 A0A084WNK2 U5EHM4 A0A023ECJ9 A0A336L9C0 B0XDC9 A0A2H1V2D6 Q16XY6 T1E2F2 Q16XY7 W8BMG1 A0A1Y1MKT1 A0A023EFU6 A0A182N2N3 A0A194RD06 A0A194PJD0 A0A1Q3FIB5 A0A182TEC5 A0A182WXW7 Q7QE07 A0A182I5Y8 A0A182FVG1 A0A2M4AGW9 T1DQL4 W5JI78 A0A182WE50 A0A182Y9M2 A0A2M4C584 A0A2M4C429 A0A182VHQ2 A0A182ISZ0 Q293S5 B4GLP6 A0A336MUQ4 A0A1W4V5A7 A0A182PM22 A0A182K180 A0A3B0JNX6 A0A0A1WG42 R4UKW7 A0A2M3ZKK1 Q9VGQ2 A0A0K8UPN9 A0A034WJ12 A0A0C9QUI6 Q8INK7 B4QID4 B3NZG3 A0A2M4B0D7 B4HMP5 A0A2A4J5S6 A0A0P9AQR0 B3LXD6 Q8SXE6 A0A1E1WDW2 A0A0Q9XA11 B4K9U8 R4WNN7 B5RIK2 A0A0Q9XD25 B4QU55 A0A1W4WUF7 A0A1W4WU50 A0A1W4WT46 A0A0J9R9N6 B4PM41 D6WIG9 A0A1E1WM56 A0A067QPA3 N6TG59 A0A212F015 Q7JW46 B4HIB1 A0A0M4EEG2 A0A0Q9WFS3 B4LXM6 A0A1J1J4G3 T1I5P0 A0A0B4KEI1 A0A1W4VQ53 A0A023FCS0 A0A0V0GBZ0 A0A0R1DMZ5 A0A1B0G4T7 A0A2P8Z1R7 A0A240SX62 A0A1B0AZE3
S4NSW2 A0A1L8E0J0 H9J1G9 A0A1B0DQY2 A0A0L7KNI9 A0A1B0CJ30 A0A0K8TRA6 B4NKV9 A0A084WNK2 U5EHM4 A0A023ECJ9 A0A336L9C0 B0XDC9 A0A2H1V2D6 Q16XY6 T1E2F2 Q16XY7 W8BMG1 A0A1Y1MKT1 A0A023EFU6 A0A182N2N3 A0A194RD06 A0A194PJD0 A0A1Q3FIB5 A0A182TEC5 A0A182WXW7 Q7QE07 A0A182I5Y8 A0A182FVG1 A0A2M4AGW9 T1DQL4 W5JI78 A0A182WE50 A0A182Y9M2 A0A2M4C584 A0A2M4C429 A0A182VHQ2 A0A182ISZ0 Q293S5 B4GLP6 A0A336MUQ4 A0A1W4V5A7 A0A182PM22 A0A182K180 A0A3B0JNX6 A0A0A1WG42 R4UKW7 A0A2M3ZKK1 Q9VGQ2 A0A0K8UPN9 A0A034WJ12 A0A0C9QUI6 Q8INK7 B4QID4 B3NZG3 A0A2M4B0D7 B4HMP5 A0A2A4J5S6 A0A0P9AQR0 B3LXD6 Q8SXE6 A0A1E1WDW2 A0A0Q9XA11 B4K9U8 R4WNN7 B5RIK2 A0A0Q9XD25 B4QU55 A0A1W4WUF7 A0A1W4WU50 A0A1W4WT46 A0A0J9R9N6 B4PM41 D6WIG9 A0A1E1WM56 A0A067QPA3 N6TG59 A0A212F015 Q7JW46 B4HIB1 A0A0M4EEG2 A0A0Q9WFS3 B4LXM6 A0A1J1J4G3 T1I5P0 A0A0B4KEI1 A0A1W4VQ53 A0A023FCS0 A0A0V0GBZ0 A0A0R1DMZ5 A0A1B0G4T7 A0A2P8Z1R7 A0A240SX62 A0A1B0AZE3
PDB
2LOM
E-value=9.7669e-11,
Score=154
Ontologies
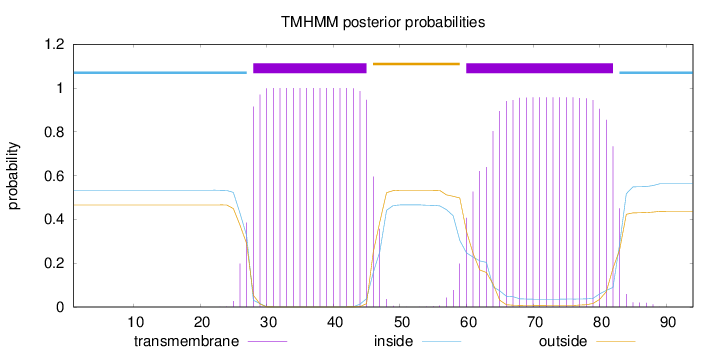
Topology
Length:
94
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.06537
Exp number, first 60 AAs:
20.15387
Total prob of N-in:
0.53362
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 45
outside
46 - 59
TMhelix
60 - 82
inside
83 - 94
Population Genetic Test Statistics
Pi
286.409361
Theta
197.172611
Tajima's D
1.370557
CLR
0
CSRT
0.754662266886656
Interpretation
Uncertain
