Gene
KWMTBOMO09187
Pre Gene Modal
BGIBMGA003359
Annotation
PREDICTED:_probable_multidrug_resistance-associated_protein_lethal(2)03659_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.019
Sequence
CDS
ATGAGGAAAAATTTGGATCCTTTTGATGAGTATTCGGACGAGGTCTTGATAAATGCACTGAAAAGTGTAGAGTTGCAAGGCGAAGGAGAGAGCGTCGGTGTACTTTACAAAGACGTGTCTGAAGGTGGATCAAACTTCAGTGTGGGAGAACGCCAACTCGTATGTTTAGCGAGAGCCATCGTCCACAACGACCGAATTTTAGTACTGGACGAGGCGACCGCTAATGTCGATGCTCAAACAGACTCCTTGATTCAAAACACCATAAGGGAACACTTCAAAGAATGCACTGTGCTTACCATTGCGCACAGATTGAACACTGTTATAGATTCTAACAAGATTTTGGTGCTAGACGCGGGTCGTCTGGTAGAGTTCGACCATCCTCATATACTTCTGCAAAACAAGAGAGGGCAATTTCGTAAAATGGTGCAGGAAACAGGCCCGTCGATGTCGAATTTACTCCACAAACTAGCCTCTCAGGTACATATCGCGATTTCTACTACAAGTAGTTATTAA
Protein
MRKNLDPFDEYSDEVLINALKSVELQGEGESVGVLYKDVSEGGSNFSVGERQLVCLARAIVHNDRILVLDEATANVDAQTDSLIQNTIREHFKECTVLTIAHRLNTVIDSNKILVLDAGRLVEFDHPHILLQNKRGQFRKMVQETGPSMSNLLHKLASQVHIAISTTSSY
Summary
Uniprot
H9J1H2
A0A437AXD7
A0A2H1W132
A0A194R8F6
A0A194PLB3
A0A2A4JL44
+ More
A0A212EZV1 A0A2H1WZW7 A0A2S2NFH7 A0A2S2P1T6 A0A2S2PMX1 X1X5I6 A0A2H8TMZ3 A0A2S2P902 A0A2S2QWL0 I4DPT5 A0A2S2N873 A0A2S2PGL8 X1XE18 A0A1B6GVC5 H9JE22 A0A2H8TFY7 J9K7F8 A0A2H8TLL8 A0A2J7PFE3 A0A2S2N6S9 A0A2J7PFD7 A0A2S2NMZ1 A0A1Y1M7A5 A0A194R8Q5 A0A2H8TSM5 A0A3Q0J0E5 A0A2S2Q6Q3 X1WJ89 A0A0T6BAF4 A0A2S2P4A1 A0A3R5SMP1 A0A0U2UWB4 A0A1W4WE54 A0A1Y1K0R4 A0A2S2RAX4 A0A2H1VX11 A0A1Y1LEY2 A0A2A4K8A3 A0A1B6MDX5 A0A067R1W1 A0A1Y1MZE9 A0A0T6AV52 A0A1Y1MZF9 A0A2S2QUD0 A0A2J7RQK1 A0A1B6H9F8 A0A0F6PRF7 A0A2S2R1S3 A0A2S2QUB8 A0A1B6JNW6 A0A1Y1K8V4 J9JY34 A0A0S7EVT1 J9JVY4 G3WRB7 H3A152 A0A1Y1LFL8 A0A1U7STS0 J9JMQ8 A0A023F3H7 A0A194PK32 A0A1Y1JYH3 A0A1Y1K348 J9K122 J9KIT1 V5G0E4 A0A161UF20 A0A0S7F0A8 A0A1B6LUH8 A0A158V0F7 A0A2J7RQI7 A0A2S2QD43 A0A3P4NGD9 A0A0P4W3Q5 A0A0K8TLH7 A0A3B4DRQ9 A0A226NZD3 A0A3B4C130 J9K2I6 A0A069DYA9 A0A1W4WP56 A0A2J7RQJ0 A0A0V0G5R1 A0A2H4KAX0 A0A0C9PVS6 A0A2R4FXD2 K7GJB7 X1X215 A0A402EI62 A0A2P8YTD5 A0A1W4WE28 A0A026VTX4 A0A3B5MW06 U4UVZ2
A0A212EZV1 A0A2H1WZW7 A0A2S2NFH7 A0A2S2P1T6 A0A2S2PMX1 X1X5I6 A0A2H8TMZ3 A0A2S2P902 A0A2S2QWL0 I4DPT5 A0A2S2N873 A0A2S2PGL8 X1XE18 A0A1B6GVC5 H9JE22 A0A2H8TFY7 J9K7F8 A0A2H8TLL8 A0A2J7PFE3 A0A2S2N6S9 A0A2J7PFD7 A0A2S2NMZ1 A0A1Y1M7A5 A0A194R8Q5 A0A2H8TSM5 A0A3Q0J0E5 A0A2S2Q6Q3 X1WJ89 A0A0T6BAF4 A0A2S2P4A1 A0A3R5SMP1 A0A0U2UWB4 A0A1W4WE54 A0A1Y1K0R4 A0A2S2RAX4 A0A2H1VX11 A0A1Y1LEY2 A0A2A4K8A3 A0A1B6MDX5 A0A067R1W1 A0A1Y1MZE9 A0A0T6AV52 A0A1Y1MZF9 A0A2S2QUD0 A0A2J7RQK1 A0A1B6H9F8 A0A0F6PRF7 A0A2S2R1S3 A0A2S2QUB8 A0A1B6JNW6 A0A1Y1K8V4 J9JY34 A0A0S7EVT1 J9JVY4 G3WRB7 H3A152 A0A1Y1LFL8 A0A1U7STS0 J9JMQ8 A0A023F3H7 A0A194PK32 A0A1Y1JYH3 A0A1Y1K348 J9K122 J9KIT1 V5G0E4 A0A161UF20 A0A0S7F0A8 A0A1B6LUH8 A0A158V0F7 A0A2J7RQI7 A0A2S2QD43 A0A3P4NGD9 A0A0P4W3Q5 A0A0K8TLH7 A0A3B4DRQ9 A0A226NZD3 A0A3B4C130 J9K2I6 A0A069DYA9 A0A1W4WP56 A0A2J7RQJ0 A0A0V0G5R1 A0A2H4KAX0 A0A0C9PVS6 A0A2R4FXD2 K7GJB7 X1X215 A0A402EI62 A0A2P8YTD5 A0A1W4WE28 A0A026VTX4 A0A3B5MW06 U4UVZ2
Pubmed
EMBL
BABH01023741
BABH01023742
BABH01023743
BABH01023744
RSAL01000296
RVE42812.1
+ More
ODYU01005653 SOQ46723.1 KQ460615 KPJ13784.1 KQ459602 KPI93524.1 NWSH01001138 PCG72438.1 AGBW02011215 OWR47028.1 ODYU01012282 SOQ58506.1 GGMR01003311 MBY15930.1 GGMR01010715 MBY23334.1 GGMR01018190 MBY30809.1 ABLF02035621 GFXV01002853 MBW14658.1 GGMR01013310 MBY25929.1 GGMS01012319 MBY81522.1 AK403773 BAM19925.1 GGMR01000367 MBY12986.1 GGMR01015946 MBY28565.1 ABLF02042979 GECZ01003398 JAS66371.1 BABH01002489 BABH01002490 GFXV01001174 MBW12979.1 ABLF02014369 GFXV01002383 MBW14188.1 NEVH01025663 PNF15053.1 GGMR01000210 MBY12829.1 PNF15046.1 GGMR01005934 MBY18553.1 GEZM01038206 JAV81719.1 KPJ13630.1 GFXV01005154 MBW16959.1 GGMS01004220 MBY73423.1 ABLF02014368 LJIG01002640 KRT84300.1 GGMR01011553 MBY24172.1 MH172501 QAA95908.1 KT276302 ALS30429.1 GEZM01099515 JAV53285.1 GGMS01018004 MBY87207.1 ODYU01004966 SOQ45370.1 GEZM01059508 JAV71448.1 NWSH01000076 PCG79892.1 GEBQ01005888 JAT34089.1 KK853083 KDR11623.1 GEZM01016939 JAV91033.1 LJIG01022813 KRT78583.1 GEZM01016941 GEZM01016940 GEZM01016938 JAV91034.1 GGMS01012154 MBY81357.1 NEVH01001336 PNF43103.1 GECU01036358 JAS71348.1 KM453743 KP335688 AKC34058.1 AKC34900.1 GGMS01014778 MBY83981.1 GGMS01011917 MBY81120.1 GECU01006806 JAT00901.1 GEZM01091336 JAV56901.1 ABLF02027734 GBYX01473114 JAO08540.1 ABLF02027727 ABLF02027732 AEFK01129726 AEFK01129727 AEFK01129728 AEFK01129729 AEFK01129730 AEFK01129731 AEFK01129732 AEFK01129733 AEFK01129734 AEFK01129735 AFYH01243657 AFYH01243658 AFYH01243659 AFYH01243660 AFYH01243661 AFYH01243662 AFYH01243663 AFYH01243664 GEZM01061568 JAV70366.1 GBBI01002866 JAC15846.1 KPI93378.1 GEZM01099513 JAV53288.1 GEZM01099514 JAV53287.1 ABLF02035628 ABLF02035624 GALX01005041 JAB63425.1 KF828787 AIN44104.1 GBYX01473128 JAO08526.1 GEBQ01012668 JAT27309.1 KF828786 AIN44103.1 PNF43104.1 GGMS01006388 MBY75591.1 CYRY02020558 VCW97093.1 GDKW01000545 JAI56050.1 GDAI01002419 JAI15184.1 AWGT02000258 OXB72712.1 ABLF02014366 GBGD01000127 JAC88762.1 PNF43102.1 GECL01002731 JAP03393.1 KY484779 ASS36021.1 GBYB01005573 JAG75340.1 MF034776 AVT42142.1 AGCU01023494 AGCU01023495 AGCU01023496 AGCU01023497 AGCU01023498 AGCU01023499 ABLF02035626 BDOT01000013 GCF43884.1 PYGN01000371 PSN47509.1 KK107928 EZA47172.1 KB632390 ERL94421.1
ODYU01005653 SOQ46723.1 KQ460615 KPJ13784.1 KQ459602 KPI93524.1 NWSH01001138 PCG72438.1 AGBW02011215 OWR47028.1 ODYU01012282 SOQ58506.1 GGMR01003311 MBY15930.1 GGMR01010715 MBY23334.1 GGMR01018190 MBY30809.1 ABLF02035621 GFXV01002853 MBW14658.1 GGMR01013310 MBY25929.1 GGMS01012319 MBY81522.1 AK403773 BAM19925.1 GGMR01000367 MBY12986.1 GGMR01015946 MBY28565.1 ABLF02042979 GECZ01003398 JAS66371.1 BABH01002489 BABH01002490 GFXV01001174 MBW12979.1 ABLF02014369 GFXV01002383 MBW14188.1 NEVH01025663 PNF15053.1 GGMR01000210 MBY12829.1 PNF15046.1 GGMR01005934 MBY18553.1 GEZM01038206 JAV81719.1 KPJ13630.1 GFXV01005154 MBW16959.1 GGMS01004220 MBY73423.1 ABLF02014368 LJIG01002640 KRT84300.1 GGMR01011553 MBY24172.1 MH172501 QAA95908.1 KT276302 ALS30429.1 GEZM01099515 JAV53285.1 GGMS01018004 MBY87207.1 ODYU01004966 SOQ45370.1 GEZM01059508 JAV71448.1 NWSH01000076 PCG79892.1 GEBQ01005888 JAT34089.1 KK853083 KDR11623.1 GEZM01016939 JAV91033.1 LJIG01022813 KRT78583.1 GEZM01016941 GEZM01016940 GEZM01016938 JAV91034.1 GGMS01012154 MBY81357.1 NEVH01001336 PNF43103.1 GECU01036358 JAS71348.1 KM453743 KP335688 AKC34058.1 AKC34900.1 GGMS01014778 MBY83981.1 GGMS01011917 MBY81120.1 GECU01006806 JAT00901.1 GEZM01091336 JAV56901.1 ABLF02027734 GBYX01473114 JAO08540.1 ABLF02027727 ABLF02027732 AEFK01129726 AEFK01129727 AEFK01129728 AEFK01129729 AEFK01129730 AEFK01129731 AEFK01129732 AEFK01129733 AEFK01129734 AEFK01129735 AFYH01243657 AFYH01243658 AFYH01243659 AFYH01243660 AFYH01243661 AFYH01243662 AFYH01243663 AFYH01243664 GEZM01061568 JAV70366.1 GBBI01002866 JAC15846.1 KPI93378.1 GEZM01099513 JAV53288.1 GEZM01099514 JAV53287.1 ABLF02035628 ABLF02035624 GALX01005041 JAB63425.1 KF828787 AIN44104.1 GBYX01473128 JAO08526.1 GEBQ01012668 JAT27309.1 KF828786 AIN44103.1 PNF43104.1 GGMS01006388 MBY75591.1 CYRY02020558 VCW97093.1 GDKW01000545 JAI56050.1 GDAI01002419 JAI15184.1 AWGT02000258 OXB72712.1 ABLF02014366 GBGD01000127 JAC88762.1 PNF43102.1 GECL01002731 JAP03393.1 KY484779 ASS36021.1 GBYB01005573 JAG75340.1 MF034776 AVT42142.1 AGCU01023494 AGCU01023495 AGCU01023496 AGCU01023497 AGCU01023498 AGCU01023499 ABLF02035626 BDOT01000013 GCF43884.1 PYGN01000371 PSN47509.1 KK107928 EZA47172.1 KB632390 ERL94421.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9J1H2
A0A437AXD7
A0A2H1W132
A0A194R8F6
A0A194PLB3
A0A2A4JL44
+ More
A0A212EZV1 A0A2H1WZW7 A0A2S2NFH7 A0A2S2P1T6 A0A2S2PMX1 X1X5I6 A0A2H8TMZ3 A0A2S2P902 A0A2S2QWL0 I4DPT5 A0A2S2N873 A0A2S2PGL8 X1XE18 A0A1B6GVC5 H9JE22 A0A2H8TFY7 J9K7F8 A0A2H8TLL8 A0A2J7PFE3 A0A2S2N6S9 A0A2J7PFD7 A0A2S2NMZ1 A0A1Y1M7A5 A0A194R8Q5 A0A2H8TSM5 A0A3Q0J0E5 A0A2S2Q6Q3 X1WJ89 A0A0T6BAF4 A0A2S2P4A1 A0A3R5SMP1 A0A0U2UWB4 A0A1W4WE54 A0A1Y1K0R4 A0A2S2RAX4 A0A2H1VX11 A0A1Y1LEY2 A0A2A4K8A3 A0A1B6MDX5 A0A067R1W1 A0A1Y1MZE9 A0A0T6AV52 A0A1Y1MZF9 A0A2S2QUD0 A0A2J7RQK1 A0A1B6H9F8 A0A0F6PRF7 A0A2S2R1S3 A0A2S2QUB8 A0A1B6JNW6 A0A1Y1K8V4 J9JY34 A0A0S7EVT1 J9JVY4 G3WRB7 H3A152 A0A1Y1LFL8 A0A1U7STS0 J9JMQ8 A0A023F3H7 A0A194PK32 A0A1Y1JYH3 A0A1Y1K348 J9K122 J9KIT1 V5G0E4 A0A161UF20 A0A0S7F0A8 A0A1B6LUH8 A0A158V0F7 A0A2J7RQI7 A0A2S2QD43 A0A3P4NGD9 A0A0P4W3Q5 A0A0K8TLH7 A0A3B4DRQ9 A0A226NZD3 A0A3B4C130 J9K2I6 A0A069DYA9 A0A1W4WP56 A0A2J7RQJ0 A0A0V0G5R1 A0A2H4KAX0 A0A0C9PVS6 A0A2R4FXD2 K7GJB7 X1X215 A0A402EI62 A0A2P8YTD5 A0A1W4WE28 A0A026VTX4 A0A3B5MW06 U4UVZ2
A0A212EZV1 A0A2H1WZW7 A0A2S2NFH7 A0A2S2P1T6 A0A2S2PMX1 X1X5I6 A0A2H8TMZ3 A0A2S2P902 A0A2S2QWL0 I4DPT5 A0A2S2N873 A0A2S2PGL8 X1XE18 A0A1B6GVC5 H9JE22 A0A2H8TFY7 J9K7F8 A0A2H8TLL8 A0A2J7PFE3 A0A2S2N6S9 A0A2J7PFD7 A0A2S2NMZ1 A0A1Y1M7A5 A0A194R8Q5 A0A2H8TSM5 A0A3Q0J0E5 A0A2S2Q6Q3 X1WJ89 A0A0T6BAF4 A0A2S2P4A1 A0A3R5SMP1 A0A0U2UWB4 A0A1W4WE54 A0A1Y1K0R4 A0A2S2RAX4 A0A2H1VX11 A0A1Y1LEY2 A0A2A4K8A3 A0A1B6MDX5 A0A067R1W1 A0A1Y1MZE9 A0A0T6AV52 A0A1Y1MZF9 A0A2S2QUD0 A0A2J7RQK1 A0A1B6H9F8 A0A0F6PRF7 A0A2S2R1S3 A0A2S2QUB8 A0A1B6JNW6 A0A1Y1K8V4 J9JY34 A0A0S7EVT1 J9JVY4 G3WRB7 H3A152 A0A1Y1LFL8 A0A1U7STS0 J9JMQ8 A0A023F3H7 A0A194PK32 A0A1Y1JYH3 A0A1Y1K348 J9K122 J9KIT1 V5G0E4 A0A161UF20 A0A0S7F0A8 A0A1B6LUH8 A0A158V0F7 A0A2J7RQI7 A0A2S2QD43 A0A3P4NGD9 A0A0P4W3Q5 A0A0K8TLH7 A0A3B4DRQ9 A0A226NZD3 A0A3B4C130 J9K2I6 A0A069DYA9 A0A1W4WP56 A0A2J7RQJ0 A0A0V0G5R1 A0A2H4KAX0 A0A0C9PVS6 A0A2R4FXD2 K7GJB7 X1X215 A0A402EI62 A0A2P8YTD5 A0A1W4WE28 A0A026VTX4 A0A3B5MW06 U4UVZ2
PDB
6BHU
E-value=3.50573e-37,
Score=383
Ontologies
GO
PANTHER
Topology
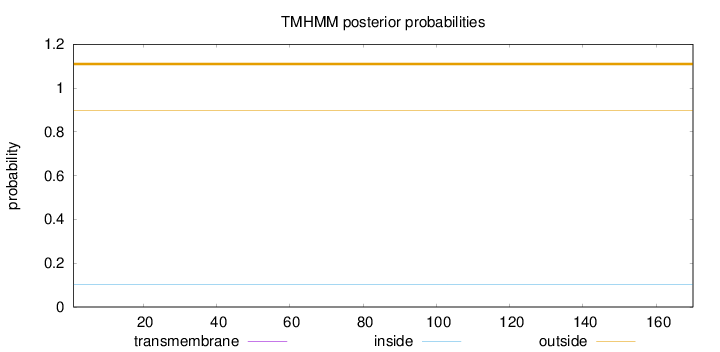
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00081
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10267
outside
1 - 170
Population Genetic Test Statistics
Pi
231.273851
Theta
181.244397
Tajima's D
0.868506
CLR
0.034426
CSRT
0.619769011549423
Interpretation
Uncertain
