Gene
KWMTBOMO09183 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003361
Annotation
Karyopherin_beta_3_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.09
Sequence
CDS
ATGGCAGGAGATCAGGCGCAATTTTATCAGTTGCTAAATACTTTACTTTCAACCGATAATGATATCAGATCCCAAGCAGAGGACGCATACAACAACATTCCAACAGAAACAAAAGTAGTGCACCTAGTGAATTCAATTCAAAATGCAGACATTGCCGAAGATGTGCGTCAAACGGCGGCCGTGCTGTTACGAAGATTATTTAGTGCAGAATTCTTTGAGTTCTTCCCGAAATTGCCATTTGAGCAGCAGACAGTGCTCAGAGAACAATTGTTACTCACCCTTCAAATGGATCTTTCCCAGTACTTACGAAGAAAGGTTTGTGATGTAGTCTCTGAACTGGCTAGGAATCATATTGATGACGATGGCAACAACCAGTGGCCTGAGTTCCTACAGTTCATGTTTACCTGTGCCAGTGCGCAAGATCCAAACATCAAAGAAGCTGGTATTAGAATGTTTACATCTGTACCAGGTGTATTTGGAAATCGTCAGACTGAAAACTTGGATGTGATAAAAGGCATGTTAATATCTGCTTTGCAGCCAAATGAATCAATGGCTTTGCGAACTCAGGCAGTTAAAGCTGTAGGCGCCTTTATACTACTACACGATAAGGAGCCCATCATACAAAAGCATTTCAGTGATGTTCTTCTACCTTTGATGCAGGTAATAGTTCTGTCTATTGAAGCAGCTGATGATGACTCTGCATTGAAAGTACTCATTGAGTTAGCAGAATCAACACCTAAATTTCTCCGACCCCAGCTGGAGACCATATTCCAAGTTTGCATAAAGGTGGTTGGGGACAAAGATGCTGAAGATAACTGGCGCCAACTGGCACTTGAAGCGATGGTAACATTATGTGAGACAGCACCAGCCATGGTGCGTAAACAAGTGCCCAATGCCGTGAGCATGCTCACTCCACTTGTACTTGAGATGATGTGTGAGCTTGATGAAGAGCCTGACTGGACCATACAGGACAACACTGCTGATGACGATAATGACCAGAATTATGTGGCAGCAGAATCAGCATTGGACCGCATGTGCTGTGGTCTTGGTGGCAAGATAATGCTGGGACTTATTGTGGGACAAGTACCTGTAATGTTGCAATCTGAAGATTGGAAGAGACGCCATGCTGCCTTGATGGCTGTCTCCTCAGCTGGTGAAGGGTGTCATAAACAGATGGAACAGATGCTCGACCAAGTTGTTACTGCTGTTCTTAATTACCTGAGTGACCCTCATCCCCGAGTACGTTATGCAGCTTGCAATGCAGTAGGGCAAATGTCAACTGATTTTGCACCAGTGTTTGAAAAAAAATTCCACAGCAAAGTTGTACCTGGACTTCTGATGGTGCTCGATGATAATGCTAACCCGCGTGTACAGGCTCATGCAGCAGCTGCTTTAGTAAACTTCAGTGAAGACTGTCCCAAACCTATCTTGACTCTGTATCTTGACCCATTAATGAAAAAGCTAGAGGCCGTCCTAACTACAAAATTTAAAGAGTTGGTTGAATGTGGTACAAAACTGGTACTCGAACAGATTGTGACAACAATCGCCTCAGTTGCAGACACTGTTGAGAAAGACTTTGTCAAATATTATGACAGACTCATGCCCTGTCTGAAGTATATAATTGCCAATGCTACTACTGATCAACTTAAGATGCTGCGTGGCAAGACAATAGAGTGTGTCAGTTTGATTGGACTAGCTGTTGGAGAAGAGAAATTCATGTCTGATGCATCTGAAGTGATGGATTTACTTCTTAAGACCCACACAGAAGGTGAACAGTTGCCTGCTGATGATCCTCAGACATCATATCTTATCTCAGCTTGGTCTAGAATTTGCAGAATAATGGGCAAGAAATTCTCACAATATTTACCGATGGTTATGGAGCCGGTAATGCGTACAGCAGCAATGAAGCCAGAAGTAGCACTTCTTGATAATGAAGATTTAGAAACCATTGAAGGAGACCTTGATTGGCATTTTGTCACACTAGGCGAACAACAAAACTTCGGCATTAAAACTGCTGGACTTGAAGACAAAGCTTCTGCTTGTGACATGTTAGTTTGCTATGCTCGAGAACTAAAGGAAGCATTTGCTGAATATGCTGAAGACGTTGTAAAGCTAATGGTGCCCATGTTGAAGTTTTATTTTCACGATAATGTGAGAACTGCTGCAGCAGAATCCCTGCCTTATCTCCTTGAATGCGCCCGCATTCGCGGCCCGCAGTATATTCAAGGAATGTGGGCTTACATAGTTCCTGAACTGCTTAAAGCCATTGACTCTGAGCCCGAACAAGAAGTGCAAGTTGAACTGCTCAACAGCTTAGCCAAATGTATCGAACTTTTGGGCATGGGTTGCCTTTCTAACAATGAGATGATGGCAGAAGTCTTGCGTATTCTCAACAAACTATTAACTGAACATTTCGAACGGGCTACTGAAAGGCGACAAAAGCGTGCCGACGAAGATTATGATGAGGTCTTAGAAGAACAATTAGCTGATGAAGATAATGAGGATGTGTACAGTTTGTCTCGCGTAGCAGATGTATTACATGCGCTCATGTCTGCTTACCGCGATTCGTTCTATCCTCATTTCGACACGTTGTTGCCCCATCTTGTGCAGCTACTTGCGCCTAATAGGCCTTATTCTGATCGCCAGTGGGCCATTTGTATTTTTGATGACATAATAGAATTCGGAGGTCCTGCTTGTGTGAAGTATCAAGATATATTCCTGGAGCCAATGTTGAATGCATTACGTGAATCTGAACCGGAAGTGCGTCAAGCCGCGGCCTACGGTTGCGGCGTGCTAGCACAGTTTGGTGACGTACAGTTTGCAGCCGCCTGCGCACGAGCTGTACCTCTTTTGGCCGCCATTGTCGCAGAGCCGGACGCACGCTCTCTTGAAAAGATCAATGCAACTGAAAATGCTATATCTGCCGTCGCTAAAATTATCAAATACAATCATTCGCAAATAAATAGGGATGAAATCATCACGCATTGGCTGACATGGTTACCGGTGACTGAAGATACCGAAGAAGCGCCTCACGTGTACGCCTTATTATGCGAGCTCGCGAGCAGCGGACACGGCGCTTTATCGACACCTGATGCCCCGCAACGTGTGCTGGCCACACTCGCCGAAGCGTTTCGCGCCGACGCTGTACCAAACGATAACCCTGTCTATGCGCAAATGGTTGCATTACTTCGACAGATACAGTCAAATGCCGAATTATTTAATTCATGTCTCATGACTTTAAGTAACGACCACAAGGAAGCTCTTCAGATAGCTCTGTCCACATAG
Protein
MAGDQAQFYQLLNTLLSTDNDIRSQAEDAYNNIPTETKVVHLVNSIQNADIAEDVRQTAAVLLRRLFSAEFFEFFPKLPFEQQTVLREQLLLTLQMDLSQYLRRKVCDVVSELARNHIDDDGNNQWPEFLQFMFTCASAQDPNIKEAGIRMFTSVPGVFGNRQTENLDVIKGMLISALQPNESMALRTQAVKAVGAFILLHDKEPIIQKHFSDVLLPLMQVIVLSIEAADDDSALKVLIELAESTPKFLRPQLETIFQVCIKVVGDKDAEDNWRQLALEAMVTLCETAPAMVRKQVPNAVSMLTPLVLEMMCELDEEPDWTIQDNTADDDNDQNYVAAESALDRMCCGLGGKIMLGLIVGQVPVMLQSEDWKRRHAALMAVSSAGEGCHKQMEQMLDQVVTAVLNYLSDPHPRVRYAACNAVGQMSTDFAPVFEKKFHSKVVPGLLMVLDDNANPRVQAHAAAALVNFSEDCPKPILTLYLDPLMKKLEAVLTTKFKELVECGTKLVLEQIVTTIASVADTVEKDFVKYYDRLMPCLKYIIANATTDQLKMLRGKTIECVSLIGLAVGEEKFMSDASEVMDLLLKTHTEGEQLPADDPQTSYLISAWSRICRIMGKKFSQYLPMVMEPVMRTAAMKPEVALLDNEDLETIEGDLDWHFVTLGEQQNFGIKTAGLEDKASACDMLVCYARELKEAFAEYAEDVVKLMVPMLKFYFHDNVRTAAAESLPYLLECARIRGPQYIQGMWAYIVPELLKAIDSEPEQEVQVELLNSLAKCIELLGMGCLSNNEMMAEVLRILNKLLTEHFERATERRQKRADEDYDEVLEEQLADEDNEDVYSLSRVADVLHALMSAYRDSFYPHFDTLLPHLVQLLAPNRPYSDRQWAICIFDDIIEFGGPACVKYQDIFLEPMLNALRESEPEVRQAAAYGCGVLAQFGDVQFAAACARAVPLLAAIVAEPDARSLEKINATENAISAVAKIIKYNHSQINRDEIITHWLTWLPVTEDTEEAPHVYALLCELASSGHGALSTPDAPQRVLATLAEAFRADAVPNDNPVYAQMVALLRQIQSNAELFNSCLMTLSNDHKEALQIALST
Summary
Uniprot
A0A437AXH0
A0A2H1V8C6
S4PY27
A0A212F003
A0A194R946
B2DBK1
+ More
A0A194PQW2 A0A1E1VZE6 A0A2A4JLX1 A0A1E1W3B5 A0A2A3E2L3 A0A0L7QTP7 A0A411G812 A0A0J7KV90 E2C279 A0A195BWU9 A0A158NCQ5 A0A151XEW1 A0A195DTB7 F4WF98 A0A067R296 A0A026WW52 A0A087ZRD4 A0A154P681 A0A2J7QL87 A0A195F8Y5 E2A245 A0A195CG91 K7IQ26 Q16TN4 A0A146LKY4 A0A0A9W6Y6 A0A1Q3F1M7 A0A1Q3F1N1 A0A182G4D0 B0WUN0 D6WJC0 A0A0V0G3A2 A0A069DY50 A0A224XH09 A0A023F3M2 A0A232F880 A0A1W4XRS7 A0A0C9R238 A0A336LZM0 A0A0P4VQV9 T1HT64 A0A182R5U2 A0A182MCE6 A0A182VXX6 A0A1Y1MPS2 A0A182P7A8 A0A182VBS7 A0A182LAS7 A0A182WV88 Q7QFZ0 A0A182I8W2 A0A182N4G8 B4KBI8 A0A182JDZ1 A0A2M4BB56 A0A0A1XKV3 T1PE61 A0A182XXG2 A0A2M3ZYV6 A0A182QXG6 W5JBT9 B3P233 A0A2M3YXF8 A0A0M5JCN0 A0A2M3ZYT8 A0A2M3ZYW0 B4LZ56 A0A182F7N6 B4PUF9 A0A182TJ41 U5EUP3 A0A1L8DXF0 B4JGW9 W8C5Y3 B4N9B5 A0A0P4WH01 A0A0N0BJH1 B3M0N4 A0A1L8DXI9 Q296D7 B4GFA9 A0A1E1WCN8 A0A034WDZ9 A0A0K8VH12 B4QVK9 A0A1W4UPI7 B4I3R5 A0A0L0CPM0 A0A3B0KBU8 Q9VN44 A0A1L8ECL5 E9H8J5 A0A0P5SV27 A0A1I8Q0B7
A0A194PQW2 A0A1E1VZE6 A0A2A4JLX1 A0A1E1W3B5 A0A2A3E2L3 A0A0L7QTP7 A0A411G812 A0A0J7KV90 E2C279 A0A195BWU9 A0A158NCQ5 A0A151XEW1 A0A195DTB7 F4WF98 A0A067R296 A0A026WW52 A0A087ZRD4 A0A154P681 A0A2J7QL87 A0A195F8Y5 E2A245 A0A195CG91 K7IQ26 Q16TN4 A0A146LKY4 A0A0A9W6Y6 A0A1Q3F1M7 A0A1Q3F1N1 A0A182G4D0 B0WUN0 D6WJC0 A0A0V0G3A2 A0A069DY50 A0A224XH09 A0A023F3M2 A0A232F880 A0A1W4XRS7 A0A0C9R238 A0A336LZM0 A0A0P4VQV9 T1HT64 A0A182R5U2 A0A182MCE6 A0A182VXX6 A0A1Y1MPS2 A0A182P7A8 A0A182VBS7 A0A182LAS7 A0A182WV88 Q7QFZ0 A0A182I8W2 A0A182N4G8 B4KBI8 A0A182JDZ1 A0A2M4BB56 A0A0A1XKV3 T1PE61 A0A182XXG2 A0A2M3ZYV6 A0A182QXG6 W5JBT9 B3P233 A0A2M3YXF8 A0A0M5JCN0 A0A2M3ZYT8 A0A2M3ZYW0 B4LZ56 A0A182F7N6 B4PUF9 A0A182TJ41 U5EUP3 A0A1L8DXF0 B4JGW9 W8C5Y3 B4N9B5 A0A0P4WH01 A0A0N0BJH1 B3M0N4 A0A1L8DXI9 Q296D7 B4GFA9 A0A1E1WCN8 A0A034WDZ9 A0A0K8VH12 B4QVK9 A0A1W4UPI7 B4I3R5 A0A0L0CPM0 A0A3B0KBU8 Q9VN44 A0A1L8ECL5 E9H8J5 A0A0P5SV27 A0A1I8Q0B7
Pubmed
23622113
22118469
26354079
18712529
20798317
21347285
+ More
21719571 24845553 24508170 30249741 20075255 17510324 26823975 25401762 26483478 18362917 19820115 26334808 25474469 28648823 27129103 28004739 20966253 12364791 14747013 17210077 17994087 25830018 25315136 25244985 20920257 23761445 17550304 24495485 15632085 25348373 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972
21719571 24845553 24508170 30249741 20075255 17510324 26823975 25401762 26483478 18362917 19820115 26334808 25474469 28648823 27129103 28004739 20966253 12364791 14747013 17210077 17994087 25830018 25315136 25244985 20920257 23761445 17550304 24495485 15632085 25348373 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972
EMBL
RSAL01000300
RVE42763.1
ODYU01001202
SOQ37098.1
GAIX01004018
JAA88542.1
+ More
AGBW02011215 OWR47024.1 KQ460615 KPJ13780.1 AB264684 BAG30760.1 KQ459602 KPI93520.1 GDQN01010961 JAT80093.1 NWSH01001138 PCG72433.1 GDQN01009600 JAT81454.1 KZ288441 PBC25714.1 KQ414740 KOC62023.1 MH366222 QBB02031.1 LBMM01002897 KMQ94211.1 GL452088 EFN77942.1 KQ976396 KYM93057.1 ADTU01012030 KQ982215 KYQ58922.1 KQ980419 KYN16088.1 GL888115 EGI67149.1 KK852751 KDR17150.1 KK107087 QOIP01000001 EZA59936.1 RLU27273.1 KQ434809 KZC06650.1 NEVH01013250 PNF29318.1 KQ981727 KYN36903.1 GL435917 EFN72493.1 KQ977791 KYM99737.1 CH477643 EAT37896.1 GDHC01011549 JAQ07080.1 GBHO01040438 GBRD01012215 JAG03166.1 JAG53609.1 GFDL01013589 JAV21456.1 GFDL01013581 JAV21464.1 JXUM01042299 KQ561318 KXJ79010.1 DS232107 EDS35015.1 KQ971343 EFA03140.1 GECL01003571 JAP02553.1 GBGD01000222 JAC88667.1 GFTR01008564 JAW07862.1 GBBI01002746 JAC15966.1 NNAY01000707 OXU26865.1 GBYB01010174 JAG79941.1 UFQT01000257 SSX22471.1 GDKW01001180 JAI55415.1 ACPB03015581 AXCM01007023 GEZM01024997 JAV87651.1 AAAB01008844 EAA06045.3 APCN01003126 CH933806 EDW13655.1 GGFJ01001144 MBW50285.1 GBXI01002323 JAD11969.1 KA647062 KA649516 AFP61691.1 GGFK01000364 MBW33685.1 AXCN02000040 ADMH02001961 ETN60244.1 CH954181 EDV47806.1 GGFM01000194 MBW20945.1 CP012526 ALC46856.1 GGFK01000344 MBW33665.1 GGFK01000341 MBW33662.1 CH940650 EDW67063.1 CM000160 EDW95681.1 GANO01002234 JAB57637.1 GFDF01002947 JAV11137.1 CH916369 EDV93747.1 GAMC01001427 JAC05129.1 CH964232 EDW80548.1 GDRN01055802 JAI65914.1 KQ435719 KOX78914.1 CH902617 EDV44281.1 GFDF01002948 JAV11136.1 CM000070 EAL28521.1 CH479182 EDW34294.1 GDQN01006335 JAT84719.1 GAKP01006587 JAC52365.1 GDHF01014494 JAI37820.1 CM000364 EDX11634.1 CH480821 EDW54858.1 JRES01000091 KNC34206.1 OUUW01000007 SPP83609.1 AE014297 BT082093 AAF52107.1 ACQ41868.1 AGB95642.1 GFDG01002375 JAV16424.1 GL732605 EFX71831.1 GDIP01137924 JAL65790.1
AGBW02011215 OWR47024.1 KQ460615 KPJ13780.1 AB264684 BAG30760.1 KQ459602 KPI93520.1 GDQN01010961 JAT80093.1 NWSH01001138 PCG72433.1 GDQN01009600 JAT81454.1 KZ288441 PBC25714.1 KQ414740 KOC62023.1 MH366222 QBB02031.1 LBMM01002897 KMQ94211.1 GL452088 EFN77942.1 KQ976396 KYM93057.1 ADTU01012030 KQ982215 KYQ58922.1 KQ980419 KYN16088.1 GL888115 EGI67149.1 KK852751 KDR17150.1 KK107087 QOIP01000001 EZA59936.1 RLU27273.1 KQ434809 KZC06650.1 NEVH01013250 PNF29318.1 KQ981727 KYN36903.1 GL435917 EFN72493.1 KQ977791 KYM99737.1 CH477643 EAT37896.1 GDHC01011549 JAQ07080.1 GBHO01040438 GBRD01012215 JAG03166.1 JAG53609.1 GFDL01013589 JAV21456.1 GFDL01013581 JAV21464.1 JXUM01042299 KQ561318 KXJ79010.1 DS232107 EDS35015.1 KQ971343 EFA03140.1 GECL01003571 JAP02553.1 GBGD01000222 JAC88667.1 GFTR01008564 JAW07862.1 GBBI01002746 JAC15966.1 NNAY01000707 OXU26865.1 GBYB01010174 JAG79941.1 UFQT01000257 SSX22471.1 GDKW01001180 JAI55415.1 ACPB03015581 AXCM01007023 GEZM01024997 JAV87651.1 AAAB01008844 EAA06045.3 APCN01003126 CH933806 EDW13655.1 GGFJ01001144 MBW50285.1 GBXI01002323 JAD11969.1 KA647062 KA649516 AFP61691.1 GGFK01000364 MBW33685.1 AXCN02000040 ADMH02001961 ETN60244.1 CH954181 EDV47806.1 GGFM01000194 MBW20945.1 CP012526 ALC46856.1 GGFK01000344 MBW33665.1 GGFK01000341 MBW33662.1 CH940650 EDW67063.1 CM000160 EDW95681.1 GANO01002234 JAB57637.1 GFDF01002947 JAV11137.1 CH916369 EDV93747.1 GAMC01001427 JAC05129.1 CH964232 EDW80548.1 GDRN01055802 JAI65914.1 KQ435719 KOX78914.1 CH902617 EDV44281.1 GFDF01002948 JAV11136.1 CM000070 EAL28521.1 CH479182 EDW34294.1 GDQN01006335 JAT84719.1 GAKP01006587 JAC52365.1 GDHF01014494 JAI37820.1 CM000364 EDX11634.1 CH480821 EDW54858.1 JRES01000091 KNC34206.1 OUUW01000007 SPP83609.1 AE014297 BT082093 AAF52107.1 ACQ41868.1 AGB95642.1 GFDG01002375 JAV16424.1 GL732605 EFX71831.1 GDIP01137924 JAL65790.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000218220
UP000242457
+ More
UP000053825 UP000036403 UP000008237 UP000078540 UP000005205 UP000075809 UP000078492 UP000007755 UP000027135 UP000053097 UP000279307 UP000005203 UP000076502 UP000235965 UP000078541 UP000000311 UP000078542 UP000002358 UP000008820 UP000069940 UP000249989 UP000002320 UP000007266 UP000215335 UP000192223 UP000015103 UP000075900 UP000075883 UP000075920 UP000075885 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075884 UP000009192 UP000075880 UP000095301 UP000076408 UP000075886 UP000000673 UP000008711 UP000092553 UP000008792 UP000069272 UP000002282 UP000075902 UP000001070 UP000007798 UP000053105 UP000007801 UP000001819 UP000008744 UP000000304 UP000192221 UP000001292 UP000037069 UP000268350 UP000000803 UP000000305 UP000095300
UP000053825 UP000036403 UP000008237 UP000078540 UP000005205 UP000075809 UP000078492 UP000007755 UP000027135 UP000053097 UP000279307 UP000005203 UP000076502 UP000235965 UP000078541 UP000000311 UP000078542 UP000002358 UP000008820 UP000069940 UP000249989 UP000002320 UP000007266 UP000215335 UP000192223 UP000015103 UP000075900 UP000075883 UP000075920 UP000075885 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075884 UP000009192 UP000075880 UP000095301 UP000076408 UP000075886 UP000000673 UP000008711 UP000092553 UP000008792 UP000069272 UP000002282 UP000075902 UP000001070 UP000007798 UP000053105 UP000007801 UP000001819 UP000008744 UP000000304 UP000192221 UP000001292 UP000037069 UP000268350 UP000000803 UP000000305 UP000095300
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A437AXH0
A0A2H1V8C6
S4PY27
A0A212F003
A0A194R946
B2DBK1
+ More
A0A194PQW2 A0A1E1VZE6 A0A2A4JLX1 A0A1E1W3B5 A0A2A3E2L3 A0A0L7QTP7 A0A411G812 A0A0J7KV90 E2C279 A0A195BWU9 A0A158NCQ5 A0A151XEW1 A0A195DTB7 F4WF98 A0A067R296 A0A026WW52 A0A087ZRD4 A0A154P681 A0A2J7QL87 A0A195F8Y5 E2A245 A0A195CG91 K7IQ26 Q16TN4 A0A146LKY4 A0A0A9W6Y6 A0A1Q3F1M7 A0A1Q3F1N1 A0A182G4D0 B0WUN0 D6WJC0 A0A0V0G3A2 A0A069DY50 A0A224XH09 A0A023F3M2 A0A232F880 A0A1W4XRS7 A0A0C9R238 A0A336LZM0 A0A0P4VQV9 T1HT64 A0A182R5U2 A0A182MCE6 A0A182VXX6 A0A1Y1MPS2 A0A182P7A8 A0A182VBS7 A0A182LAS7 A0A182WV88 Q7QFZ0 A0A182I8W2 A0A182N4G8 B4KBI8 A0A182JDZ1 A0A2M4BB56 A0A0A1XKV3 T1PE61 A0A182XXG2 A0A2M3ZYV6 A0A182QXG6 W5JBT9 B3P233 A0A2M3YXF8 A0A0M5JCN0 A0A2M3ZYT8 A0A2M3ZYW0 B4LZ56 A0A182F7N6 B4PUF9 A0A182TJ41 U5EUP3 A0A1L8DXF0 B4JGW9 W8C5Y3 B4N9B5 A0A0P4WH01 A0A0N0BJH1 B3M0N4 A0A1L8DXI9 Q296D7 B4GFA9 A0A1E1WCN8 A0A034WDZ9 A0A0K8VH12 B4QVK9 A0A1W4UPI7 B4I3R5 A0A0L0CPM0 A0A3B0KBU8 Q9VN44 A0A1L8ECL5 E9H8J5 A0A0P5SV27 A0A1I8Q0B7
A0A194PQW2 A0A1E1VZE6 A0A2A4JLX1 A0A1E1W3B5 A0A2A3E2L3 A0A0L7QTP7 A0A411G812 A0A0J7KV90 E2C279 A0A195BWU9 A0A158NCQ5 A0A151XEW1 A0A195DTB7 F4WF98 A0A067R296 A0A026WW52 A0A087ZRD4 A0A154P681 A0A2J7QL87 A0A195F8Y5 E2A245 A0A195CG91 K7IQ26 Q16TN4 A0A146LKY4 A0A0A9W6Y6 A0A1Q3F1M7 A0A1Q3F1N1 A0A182G4D0 B0WUN0 D6WJC0 A0A0V0G3A2 A0A069DY50 A0A224XH09 A0A023F3M2 A0A232F880 A0A1W4XRS7 A0A0C9R238 A0A336LZM0 A0A0P4VQV9 T1HT64 A0A182R5U2 A0A182MCE6 A0A182VXX6 A0A1Y1MPS2 A0A182P7A8 A0A182VBS7 A0A182LAS7 A0A182WV88 Q7QFZ0 A0A182I8W2 A0A182N4G8 B4KBI8 A0A182JDZ1 A0A2M4BB56 A0A0A1XKV3 T1PE61 A0A182XXG2 A0A2M3ZYV6 A0A182QXG6 W5JBT9 B3P233 A0A2M3YXF8 A0A0M5JCN0 A0A2M3ZYT8 A0A2M3ZYW0 B4LZ56 A0A182F7N6 B4PUF9 A0A182TJ41 U5EUP3 A0A1L8DXF0 B4JGW9 W8C5Y3 B4N9B5 A0A0P4WH01 A0A0N0BJH1 B3M0N4 A0A1L8DXI9 Q296D7 B4GFA9 A0A1E1WCN8 A0A034WDZ9 A0A0K8VH12 B4QVK9 A0A1W4UPI7 B4I3R5 A0A0L0CPM0 A0A3B0KBU8 Q9VN44 A0A1L8ECL5 E9H8J5 A0A0P5SV27 A0A1I8Q0B7
PDB
3W3Z
E-value=3.52136e-111,
Score=1031
Ontologies
GO
PANTHER
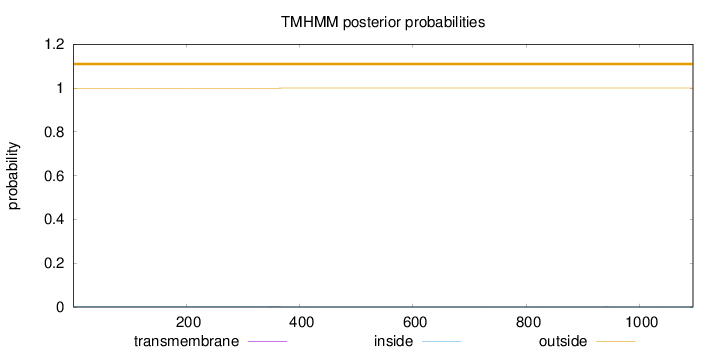
Topology
Length:
1094
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01781
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00076
outside
1 - 1094
Population Genetic Test Statistics
Pi
27.218103
Theta
218.712429
Tajima's D
0.246413
CLR
1.543441
CSRT
0.437678116094195
Interpretation
Uncertain
