Pre Gene Modal
BGIBMGA003390
Annotation
PREDICTED:_uncharacterized_protein_LOC101740766_[Bombyx_mori]
Full name
N-acetyltransferase 9
Alternative Name
Embryo brain-specific protein
Location in the cell
Cytoplasmic Reliability : 2.753
Sequence
CDS
ATGAAATTAAATAGTCATACCAAAATCGTAGGAGCAAATATAATTTTGGTTCCTTACAAAAAACATCATGTACCTGTCTATCATGAATGGATGAAGTCCGAAGAATTACAAAAGCTAACAGCTTCAGAACCATTAAATTTAGAACAAGAGTACGAAATGCAAAAAACATGGCATGATGACGAAGATAAATGCACATTTATCATTTTGGACAAGGATATATTTAATAATACAGCCTTCGATGAAACAGGTGCAATGATTGGGGATACAAATATATTCATAACTGACAAAGAGTTGGCTATAGGAGAAATAGAAATAATGATAGCTGAACCATCAGCCAGAGGAAAGAAAAGAGGATGGGAAGCTGTTAAATTAATGTTACTTTATGGAATAAAGTATATAAAAATACAAACATTTGAAGCAAAAATATTATTACAGAATGTGATCAGTATTAAAATGTTCAAAAAGTTAGGATTTCAGGAGAAAAGTGTCAGTGCAGTATTTCAAGAAGTGACTTTGGACAAGAAAGTATCAAATGAATGGACTGAGTGGTTAGCAAGTAGTCTTTCTTTTGAAATAAAAGACTATCACTAA
Protein
MKLNSHTKIVGANIILVPYKKHHVPVYHEWMKSEELQKLTASEPLNLEQEYEMQKTWHDDEDKCTFIILDKDIFNNTAFDETGAMIGDTNIFITDKELAIGEIEIMIAEPSARGKKRGWEAVKLMLLYGIKYIKIQTFEAKILLQNVISIKMFKKLGFQEKSVSAVFQEVTLDKKVSNEWTEWLASSLSFEIKDYH
Summary
Similarity
Belongs to the acetyltransferase family. GNAT subfamily.
Keywords
Acyltransferase
Alternative splicing
Complete proteome
Polymorphism
Reference proteome
Transferase
Feature
chain N-acetyltransferase 9
splice variant In isoform 2.
sequence variant In dbSNP:rs2305213.
splice variant In isoform 2.
sequence variant In dbSNP:rs2305213.
Uniprot
H9J1K3
A0A2A4ISG4
A0A2H1WHQ4
A0A0L7LG05
A0A1E1WJX1
S4P267
+ More
A0A194PJB5 A0A212EZZ0 A0A437AX59 A0A1E1VZW6 A0A1B0B386 A0A182HAW9 A0A067R6E7 A0A023EI77 A0A1A9XZ65 A0A1B0GB54 E9JB66 A0A026WVM5 F4X3D0 A0A2J7Q7X9 K7IQY6 A0A1A9ZXS6 A0A1Q3FGR7 A0A1A9VEW5 A0A158NI78 B0XIK9 Q0IEU4 A0A1S4FIB2 A0A1L8EMS5 E2C8N5 A0A336KZZ8 H3B1C3 W5JIU4 A0A1D5PI17 A0A182XYP8 A0A2P4T7K2 A0A1C7CZI3 A0A453Z3Q6 Q7QJA1 A0A3B5KC73 W5MZA3 W5MZA9 A0A0L7QS54 A0A3Q3KSG0 A0A1V4KW03 W5U8U0 A0A3Q3L478 A0A3Q3CN79 A0A444U786 A0A154P0Q1 U3ECW1 A0A3Q4MGU2 A0A3Q2QJ55 H2NUM7 G1KE42 A0A212D7T4 A0A452G6E3 A0A3P8RDC9 A0A3P9B7H4 A0A2K5NH04 F6TPU6 A0A2I4B4C7 A0A1A8BLP1 A0A1A8B1U9 L8HVS9 A0A2K5YQC8 A0A182FDE5 Q2I153 A0A3P8Z895 J3QRL7 A0A2K6E450 H9FSS4 A0A3P9B7Q2 Q9BTE0-2 A0A3Q2E7E6 A0A2K5RZX8 A0A1A7WYU8 A0A1A8RXS5 A0A1A8PRB0 A0A1A8HWV6 A0A1A8GG88 A0A2J8Y0A2 A0A1S3GYW1 A0A2K5RZV3 H0UVU9 A0A2J8Y050 A0A0L0CKJ7 A0A2J8KBW9 A0A3Q3X8P0 A0A2R9C0E1 A0A2J8KBT7 A0A151N3B5 A0A218UV48 K7FCU7 A0A3Q1HLT3 A0A1Y1MA28 V9L5H0 I3JMK6 A0A182K2A7 A0A3B3USF1
A0A194PJB5 A0A212EZZ0 A0A437AX59 A0A1E1VZW6 A0A1B0B386 A0A182HAW9 A0A067R6E7 A0A023EI77 A0A1A9XZ65 A0A1B0GB54 E9JB66 A0A026WVM5 F4X3D0 A0A2J7Q7X9 K7IQY6 A0A1A9ZXS6 A0A1Q3FGR7 A0A1A9VEW5 A0A158NI78 B0XIK9 Q0IEU4 A0A1S4FIB2 A0A1L8EMS5 E2C8N5 A0A336KZZ8 H3B1C3 W5JIU4 A0A1D5PI17 A0A182XYP8 A0A2P4T7K2 A0A1C7CZI3 A0A453Z3Q6 Q7QJA1 A0A3B5KC73 W5MZA3 W5MZA9 A0A0L7QS54 A0A3Q3KSG0 A0A1V4KW03 W5U8U0 A0A3Q3L478 A0A3Q3CN79 A0A444U786 A0A154P0Q1 U3ECW1 A0A3Q4MGU2 A0A3Q2QJ55 H2NUM7 G1KE42 A0A212D7T4 A0A452G6E3 A0A3P8RDC9 A0A3P9B7H4 A0A2K5NH04 F6TPU6 A0A2I4B4C7 A0A1A8BLP1 A0A1A8B1U9 L8HVS9 A0A2K5YQC8 A0A182FDE5 Q2I153 A0A3P8Z895 J3QRL7 A0A2K6E450 H9FSS4 A0A3P9B7Q2 Q9BTE0-2 A0A3Q2E7E6 A0A2K5RZX8 A0A1A7WYU8 A0A1A8RXS5 A0A1A8PRB0 A0A1A8HWV6 A0A1A8GG88 A0A2J8Y0A2 A0A1S3GYW1 A0A2K5RZV3 H0UVU9 A0A2J8Y050 A0A0L0CKJ7 A0A2J8KBW9 A0A3Q3X8P0 A0A2R9C0E1 A0A2J8KBT7 A0A151N3B5 A0A218UV48 K7FCU7 A0A3Q1HLT3 A0A1Y1MA28 V9L5H0 I3JMK6 A0A182K2A7 A0A3B3USF1
EC Number
2.3.1.-
Pubmed
19121390
26227816
23622113
26354079
22118469
26483478
+ More
24845553 24945155 21282665 24508170 30249741 21719571 20075255 21347285 17510324 27762356 20798317 9215903 20920257 23761445 15592404 25244985 20966253 12364791 14747013 17210077 21551351 23127152 25243066 25186727 17431167 22751099 25069045 16625196 25319552 14702039 17974005 15489334 21993624 26108605 22722832 22293439 17381049 28004739 24402279
24845553 24945155 21282665 24508170 30249741 21719571 20075255 21347285 17510324 27762356 20798317 9215903 20920257 23761445 15592404 25244985 20966253 12364791 14747013 17210077 21551351 23127152 25243066 25186727 17431167 22751099 25069045 16625196 25319552 14702039 17974005 15489334 21993624 26108605 22722832 22293439 17381049 28004739 24402279
EMBL
BABH01023729
BABH01023730
NWSH01008300
PCG62609.1
ODYU01008651
SOQ52422.1
+ More
JTDY01001255 KOB74382.1 GDQN01003762 JAT87292.1 GAIX01006964 JAA85596.1 KQ459602 KPI93516.1 AGBW02011215 OWR47021.1 RSAL01000300 RVE42760.1 GDQN01010853 JAT80201.1 JXJN01007779 JXUM01031575 KQ560926 KXJ80321.1 KK852855 KDR14922.1 GAPW01004505 JAC09093.1 CCAG010019322 GL770938 EFZ09935.1 KK107109 QOIP01000008 EZA59139.1 RLU19796.1 GL888609 EGI59111.1 NEVH01017000 PNF24691.1 GFDL01008295 JAV26750.1 ADTU01016068 DS233328 EDS29433.1 CH477478 EAT40257.1 CM004483 OCT60653.1 GL453666 EFN75765.1 UFQS01001373 UFQT01001373 SSX10555.1 SSX30239.1 AFYH01091829 ADMH02001280 ETN63213.1 AADN05000628 PPHD01006061 POI32348.1 AAAB01008807 EAA04632.3 AHAT01000258 KQ414758 KOC61475.1 LSYS01001520 OPJ88634.1 JT408534 AHH38368.1 SCEB01215145 RXM31047.1 KQ434783 KZC04720.1 GAMR01010516 JAB23416.1 ABGA01168077 MKHE01000005 OWK14154.1 LWLT01000022 JSUE03017403 HADZ01004596 HAEA01015287 SBP68537.1 HADY01022769 HAEJ01010195 SBP61254.1 JH883080 ELR47429.1 DQ353801 ABC75592.1 AC016888 JU333928 JV046722 AFE77683.1 AFI36793.1 AY632082 AK312958 AL050269 CH471099 BC004195 BC004225 HADW01009475 HADX01014996 SBP10875.1 HAEI01003706 HAEH01020258 SBS10512.1 HAEF01001564 HAEG01009067 SBR83524.1 HAED01002153 HAEE01016254 SBQ87998.1 HAEB01019039 HAEC01001969 SBQ70046.1 NDHI03003284 PNJ87668.1 AAKN02047289 PNJ87665.1 JRES01000266 KNC32790.1 NBAG03000379 PNI32500.1 AJFE02042360 AJFE02042361 PNI32494.1 AKHW03004113 KYO31115.1 MUZQ01000120 OWK57635.1 AGCU01174000 GEZM01039823 JAV81155.1 JW874316 AFP06833.1 AERX01030862
JTDY01001255 KOB74382.1 GDQN01003762 JAT87292.1 GAIX01006964 JAA85596.1 KQ459602 KPI93516.1 AGBW02011215 OWR47021.1 RSAL01000300 RVE42760.1 GDQN01010853 JAT80201.1 JXJN01007779 JXUM01031575 KQ560926 KXJ80321.1 KK852855 KDR14922.1 GAPW01004505 JAC09093.1 CCAG010019322 GL770938 EFZ09935.1 KK107109 QOIP01000008 EZA59139.1 RLU19796.1 GL888609 EGI59111.1 NEVH01017000 PNF24691.1 GFDL01008295 JAV26750.1 ADTU01016068 DS233328 EDS29433.1 CH477478 EAT40257.1 CM004483 OCT60653.1 GL453666 EFN75765.1 UFQS01001373 UFQT01001373 SSX10555.1 SSX30239.1 AFYH01091829 ADMH02001280 ETN63213.1 AADN05000628 PPHD01006061 POI32348.1 AAAB01008807 EAA04632.3 AHAT01000258 KQ414758 KOC61475.1 LSYS01001520 OPJ88634.1 JT408534 AHH38368.1 SCEB01215145 RXM31047.1 KQ434783 KZC04720.1 GAMR01010516 JAB23416.1 ABGA01168077 MKHE01000005 OWK14154.1 LWLT01000022 JSUE03017403 HADZ01004596 HAEA01015287 SBP68537.1 HADY01022769 HAEJ01010195 SBP61254.1 JH883080 ELR47429.1 DQ353801 ABC75592.1 AC016888 JU333928 JV046722 AFE77683.1 AFI36793.1 AY632082 AK312958 AL050269 CH471099 BC004195 BC004225 HADW01009475 HADX01014996 SBP10875.1 HAEI01003706 HAEH01020258 SBS10512.1 HAEF01001564 HAEG01009067 SBR83524.1 HAED01002153 HAEE01016254 SBQ87998.1 HAEB01019039 HAEC01001969 SBQ70046.1 NDHI03003284 PNJ87668.1 AAKN02047289 PNJ87665.1 JRES01000266 KNC32790.1 NBAG03000379 PNI32500.1 AJFE02042360 AJFE02042361 PNI32494.1 AKHW03004113 KYO31115.1 MUZQ01000120 OWK57635.1 AGCU01174000 GEZM01039823 JAV81155.1 JW874316 AFP06833.1 AERX01030862
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000283053
+ More
UP000092460 UP000069940 UP000249989 UP000027135 UP000092443 UP000092444 UP000053097 UP000279307 UP000007755 UP000235965 UP000002358 UP000092445 UP000078200 UP000005205 UP000002320 UP000008820 UP000186698 UP000008237 UP000008672 UP000000673 UP000000539 UP000076408 UP000075882 UP000076407 UP000007062 UP000005226 UP000018468 UP000053825 UP000261640 UP000190648 UP000221080 UP000264840 UP000076502 UP000261580 UP000265000 UP000001595 UP000001646 UP000291000 UP000265100 UP000265160 UP000233060 UP000006718 UP000192220 UP000233140 UP000069272 UP000265140 UP000005640 UP000233120 UP000265020 UP000233040 UP000081671 UP000005447 UP000037069 UP000261620 UP000240080 UP000050525 UP000197619 UP000007267 UP000265040 UP000005207 UP000075881 UP000261500
UP000092460 UP000069940 UP000249989 UP000027135 UP000092443 UP000092444 UP000053097 UP000279307 UP000007755 UP000235965 UP000002358 UP000092445 UP000078200 UP000005205 UP000002320 UP000008820 UP000186698 UP000008237 UP000008672 UP000000673 UP000000539 UP000076408 UP000075882 UP000076407 UP000007062 UP000005226 UP000018468 UP000053825 UP000261640 UP000190648 UP000221080 UP000264840 UP000076502 UP000261580 UP000265000 UP000001595 UP000001646 UP000291000 UP000265100 UP000265160 UP000233060 UP000006718 UP000192220 UP000233140 UP000069272 UP000265140 UP000005640 UP000233120 UP000265020 UP000233040 UP000081671 UP000005447 UP000037069 UP000261620 UP000240080 UP000050525 UP000197619 UP000007267 UP000265040 UP000005207 UP000075881 UP000261500
Interpro
Gene 3D
ProteinModelPortal
H9J1K3
A0A2A4ISG4
A0A2H1WHQ4
A0A0L7LG05
A0A1E1WJX1
S4P267
+ More
A0A194PJB5 A0A212EZZ0 A0A437AX59 A0A1E1VZW6 A0A1B0B386 A0A182HAW9 A0A067R6E7 A0A023EI77 A0A1A9XZ65 A0A1B0GB54 E9JB66 A0A026WVM5 F4X3D0 A0A2J7Q7X9 K7IQY6 A0A1A9ZXS6 A0A1Q3FGR7 A0A1A9VEW5 A0A158NI78 B0XIK9 Q0IEU4 A0A1S4FIB2 A0A1L8EMS5 E2C8N5 A0A336KZZ8 H3B1C3 W5JIU4 A0A1D5PI17 A0A182XYP8 A0A2P4T7K2 A0A1C7CZI3 A0A453Z3Q6 Q7QJA1 A0A3B5KC73 W5MZA3 W5MZA9 A0A0L7QS54 A0A3Q3KSG0 A0A1V4KW03 W5U8U0 A0A3Q3L478 A0A3Q3CN79 A0A444U786 A0A154P0Q1 U3ECW1 A0A3Q4MGU2 A0A3Q2QJ55 H2NUM7 G1KE42 A0A212D7T4 A0A452G6E3 A0A3P8RDC9 A0A3P9B7H4 A0A2K5NH04 F6TPU6 A0A2I4B4C7 A0A1A8BLP1 A0A1A8B1U9 L8HVS9 A0A2K5YQC8 A0A182FDE5 Q2I153 A0A3P8Z895 J3QRL7 A0A2K6E450 H9FSS4 A0A3P9B7Q2 Q9BTE0-2 A0A3Q2E7E6 A0A2K5RZX8 A0A1A7WYU8 A0A1A8RXS5 A0A1A8PRB0 A0A1A8HWV6 A0A1A8GG88 A0A2J8Y0A2 A0A1S3GYW1 A0A2K5RZV3 H0UVU9 A0A2J8Y050 A0A0L0CKJ7 A0A2J8KBW9 A0A3Q3X8P0 A0A2R9C0E1 A0A2J8KBT7 A0A151N3B5 A0A218UV48 K7FCU7 A0A3Q1HLT3 A0A1Y1MA28 V9L5H0 I3JMK6 A0A182K2A7 A0A3B3USF1
A0A194PJB5 A0A212EZZ0 A0A437AX59 A0A1E1VZW6 A0A1B0B386 A0A182HAW9 A0A067R6E7 A0A023EI77 A0A1A9XZ65 A0A1B0GB54 E9JB66 A0A026WVM5 F4X3D0 A0A2J7Q7X9 K7IQY6 A0A1A9ZXS6 A0A1Q3FGR7 A0A1A9VEW5 A0A158NI78 B0XIK9 Q0IEU4 A0A1S4FIB2 A0A1L8EMS5 E2C8N5 A0A336KZZ8 H3B1C3 W5JIU4 A0A1D5PI17 A0A182XYP8 A0A2P4T7K2 A0A1C7CZI3 A0A453Z3Q6 Q7QJA1 A0A3B5KC73 W5MZA3 W5MZA9 A0A0L7QS54 A0A3Q3KSG0 A0A1V4KW03 W5U8U0 A0A3Q3L478 A0A3Q3CN79 A0A444U786 A0A154P0Q1 U3ECW1 A0A3Q4MGU2 A0A3Q2QJ55 H2NUM7 G1KE42 A0A212D7T4 A0A452G6E3 A0A3P8RDC9 A0A3P9B7H4 A0A2K5NH04 F6TPU6 A0A2I4B4C7 A0A1A8BLP1 A0A1A8B1U9 L8HVS9 A0A2K5YQC8 A0A182FDE5 Q2I153 A0A3P8Z895 J3QRL7 A0A2K6E450 H9FSS4 A0A3P9B7Q2 Q9BTE0-2 A0A3Q2E7E6 A0A2K5RZX8 A0A1A7WYU8 A0A1A8RXS5 A0A1A8PRB0 A0A1A8HWV6 A0A1A8GG88 A0A2J8Y0A2 A0A1S3GYW1 A0A2K5RZV3 H0UVU9 A0A2J8Y050 A0A0L0CKJ7 A0A2J8KBW9 A0A3Q3X8P0 A0A2R9C0E1 A0A2J8KBT7 A0A151N3B5 A0A218UV48 K7FCU7 A0A3Q1HLT3 A0A1Y1MA28 V9L5H0 I3JMK6 A0A182K2A7 A0A3B3USF1
PDB
3EO4
E-value=0.0021312,
Score=93
Ontologies
GO
PANTHER
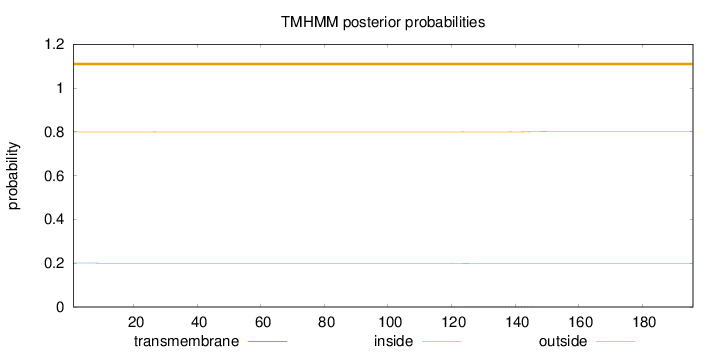
Topology
Length:
196
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05031
Exp number, first 60 AAs:
0.00222
Total prob of N-in:
0.20043
outside
1 - 196
Population Genetic Test Statistics
Pi
30.026381
Theta
29.382114
Tajima's D
0.300221
CLR
0.309124
CSRT
0.455427228638568
Interpretation
Uncertain
