Gene
KWMTBOMO09178
Pre Gene Modal
BGIBMGA003388
Annotation
PREDICTED:_lysosomal_acid_phosphatase-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.1
Sequence
CDS
ATGCAATGCGTCGATAAAACGGACCCACCCCCCACAGAGCCATTATTTCCTCAACCTACCTCAGTATGCACAGTAGCCATAAAACTTCCTCCCTTCTGGCCTGATCGGCCTGCAATTTGGTTTGCTCAGGTAGAGGCACAATTCTCGATTTCGGGCATCTCCGCTGACCTGACTAAATTTAACTACGTGATTGCTCAGTTAGATACCCGAGTTATTGGGGAGGTAGAAGACATTATTCTCCAACCACCACCAGAGGACAAATATGGTCGCCTCAAATCCGAGTTGATCCGTAGGTTGTCCACGTCTGAGGAGCAACGTGTCAGGCAACTTGTCAGTGATGTTGAGTTGGGTGACCGCCGTCCTTCGCAGTTTTTACGTCATCTTAGATCATTGGCAGGAAACACCCTCACTGATGAAAATTTGCTTCGCCAGCTATGGATGCGTCGCCTCCCACAACATCTCCAAGCAATTTTGGCCGCCCAATCAGAACTCTCGCTGGACAAAGTCGCTGAGCTAGCGGACAAAATTTTGGAAGTTTCGCCTGAAGCAACATCTAAACAAATGGAATATGATAATGATGATAACTCATGGAGAAGACCGTCCACCACTCAGCAGAATAAATGTCATCGGATTCCAAAGAGGTCGACCACAACTGCAGTAGTAGTGCTGGGCTTGGCGGTACTGAGCTGTCTCCTTGGATACTGCGTTTTGAGCGAAACATTACCTTATGAATCTAAAACTCTTCGTTTGGTTATTATCTTGTTTCGTCATGGTGCCCGAACTCCGGTTTCAACATATAGAAGTGATCCCTACAGGAACTACCAGTGGCCCGATGGATATGGTAGTCTTACAAATGTGGGAAAGATGCAATTGTACGAATTAGGAAAGAAATACCGCAGCTACTATGCCAACTACATACCCGAGGACTATTATGAGAAAAACGTGTACGTACAGAGCAGCGACAAGTCTCGTTGCATGATGAGTGCGTACACATTCCTGGCGGGACTATTCCCTCCGTCGCAACGGCAAACTTGGCATGCGGAGTTGCCATGGCAACCAATACCGGTCCACTCACTGCCCCGGCTTCTTGACAATATAGTGGCAGGCACGAAGCCTTGTGCGAAATGGAAAGCCATGTACGCGGAGCAGCTAGCTGAGAAAAATTCCGATCCGAAATTCAATGAGGTATTCGAGAATTTATCGAAATACACGAACCAGACGGTCCGCAGTGTTCTCGACGTCGACTACTTGTACAGCACCTTGGCCACGGAACAGGAAGCTGGCCTCAAACTTCCGGAATGGACAAAAAATTTGTTTCCTCACAAAATGAGACTACCCTTCATGCTGAGTTTAGCTTTGCTTTCTTACAACGAGTCCATGCAACGGCTTCAAATAGGTCCGATGCTCAACGAGATCCGTGAGAACATGAACAATACGTTTAATCCAAGCTCTGGGTCGGAACGGTTGCTGCTGATGTACTCGGGGCACGACGTGACCGTGGTGGCCTTGTGGAGGGCGCTCGGCTTCGGCGAGCTGCTGGAGCCGGAGTACGGAGCCAGCCTCGTACTCGAACTGCACGAGGACATCGAACAGAATCTCTTCTTCGTTAAGGCTTTCTATCGTAACAATACTAAAACAGAGATGCCGATGGAGCTAAAGCTGCCGTTCTGCGAAGATCCATGTGGTTATAAACAATTTTACGACCACATAAGCAACCTGATTCCTGATGATTGGGAAACTGAGTGTGAGAATAATGTTCAAGGTGCATAA
Protein
MQCVDKTDPPPTEPLFPQPTSVCTVAIKLPPFWPDRPAIWFAQVEAQFSISGISADLTKFNYVIAQLDTRVIGEVEDIILQPPPEDKYGRLKSELIRRLSTSEEQRVRQLVSDVELGDRRPSQFLRHLRSLAGNTLTDENLLRQLWMRRLPQHLQAILAAQSELSLDKVAELADKILEVSPEATSKQMEYDNDDNSWRRPSTTQQNKCHRIPKRSTTTAVVVLGLAVLSCLLGYCVLSETLPYESKTLRLVIILFRHGARTPVSTYRSDPYRNYQWPDGYGSLTNVGKMQLYELGKKYRSYYANYIPEDYYEKNVYVQSSDKSRCMMSAYTFLAGLFPPSQRQTWHAELPWQPIPVHSLPRLLDNIVAGTKPCAKWKAMYAEQLAEKNSDPKFNEVFENLSKYTNQTVRSVLDVDYLYSTLATEQEAGLKLPEWTKNLFPHKMRLPFMLSLALLSYNESMQRLQIGPMLNEIRENMNNTFNPSSGSERLLLMYSGHDVTVVALWRALGFGELLEPEYGASLVLELHEDIEQNLFFVKAFYRNNTKTEMPMELKLPFCEDPCGYKQFYDHISNLIPDDWETECENNVQGA
Summary
Uniprot
ProteinModelPortal
PDB
1RPT
E-value=1.20579e-45,
Score=463
Ontologies
PATHWAY
GO
Topology
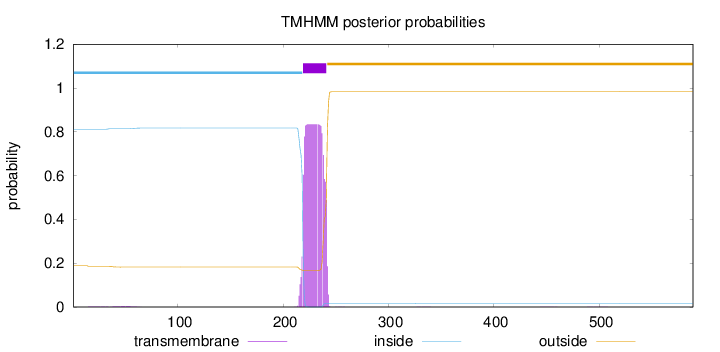
Length:
589
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.7455599999999
Exp number, first 60 AAs:
0.11604
Total prob of N-in:
0.81174
inside
1 - 218
TMhelix
219 - 241
outside
242 - 589
Population Genetic Test Statistics
Pi
234.207647
Theta
170.351902
Tajima's D
0.40381
CLR
1.231069
CSRT
0.484525773711314
Interpretation
Uncertain
