Pre Gene Modal
BGIBMGA003365
Annotation
PREDICTED:_integrator_complex_subunit_4_[Amyelois_transitella]
Full name
Integrator complex subunit 4
Location in the cell
Cytoplasmic Reliability : 1.43 Golgi Reliability : 1.324 Nuclear Reliability : 1.005
Sequence
CDS
ATGGCTGCAGTTTTAAAGAAGAGAGCCCTGGCAGAATACAATAAATCATTTCAGGATGCTCCACCATCGAAGAAGCTGCATTTGGCAAAGAAGCCGTTAATAGGTAGTTCTGCTGCTGCCTTTGTGGGTCTTCTAGAGAAATGCAAGTCCAGCGATGAAGCGTTGCAGTTTTTACTCAGAATATCCGACTGTCTTCAGTTCCAAGAGTCCGACGTAGAGGAAGCAATAAAGAAGTTGTCAAAACATTTTCAGTCGGAAGAAGAGTCTGTAGTCAGAGTAAAAATACTATGGCTTTTTTGTGACATAGGCTTGGAGGTTCCCTGTGCAAACTTGAATAACTTGATAGATGAAACCATACATTTAATAAAAAACGAAACGTCACACAAGGTTATCGGTCAAGGAATAGCAACACTGACAAAACTTGCCAAGAAATTGAGTGACGATAAAATTTTAATGATGAGGCTAGTTGGTGTTGCTAAAGACAATTTGAAAGATACTAGCCATCAGGTGAAATGTAGGTGTCTACAGCTCATCAGTGAGTTGTACCCTATACATCCGGACTCTGATAGAACATCTGAGATGACAGCAGAGGCTGAAGCTATTGTAAAACTTCTTGGAGATTATTCTAATTCTGAGGATGCCAGAGTGCGTTGTGAAGCATTTCAAGCACTGTTAACACTTCATCAAAGAGGACAGATCTTAGGATCTAATTTGTACGAACATGTTTGTGTTGCTCTGTCCGATGACTATGAAATAGTAAGAGAAGTTGCCCTAAAGTTGATTTGGGTACTTGGAAATAAATACCCAGAAAATGTAATAACGTTACAAGATGACGAGACCACGATACGAATGGTCGATGACGCTTTTGTTCGCATATGTTCGGCCGTGAACGATCTATGCATGCAAGTGCGAGCGCTCGCCTGCACGCTGCTCGGCACCACGCGCGCCGTTTCCGATCGCTTCCTGTTGCAGACCCTCGATAAACAACTAATGAGTAACATGAAGAAAAAGCGCACAGCACACGAGCGCGGCGCGGAGCTGGTCCGCAGCGGCGCGTGGGCGAGCGGGCGGCGCTGGGCGGACGACGCGCCCGGCGCGCTGGTGCCGTCCGCCGCCGTGCAGCTGCTGCCGGGCGGGGCGGCCGGCGCCTTTGTGCACGGACTCGAGGACGAACTCATGGAGGTGCGCACGGCAGCCGTCGAAGCATTGTGTCAACTCTCACTAGAAAACCCGGTGTTCGCGACCACCTCCCTCGACTTTTTGGTGGACATGTTCAATGACGAGATCGAGGAAGTGCGGCTGCGAGCCATCGACGCCCTCACGAGGATCTCGCATCATATCGTACTTAGAGAAGACCTACTGGAGACCATACTAGGAGCCTTGGAGGATTATTCAATGGACGTCAGGGAAGGTCTACACAGACTGCTGGGCTCGTGTACAGTCGCGTCCAAAACATGCCTGGAGATGTGCATTGATAAAATTCTGGAGAATCTCAAAAGATATCCACAGGATAAGCGTTCGACATTCCGTTGCGTGCAGCGGCTGGGCAGCTCGCACGCGACCCTGGTGCTGCCGCTGACCACGAGGCTGCTGGCCGTGCACCCTTTCTTCGACATGCCTGAGCCAGATGTTGAGGATCCAGCTTATATGTGCGTTTTGATCCTGGTGTTGAACGCGGCGCAGCACTGCACCACCATGCTGCCGCTGTTCGAGGAGCACACCATCAAGCATTACACGTATCTGCGGGACACCATGCCTCATCTCGTGCCGCATCTGCCCATCGGCGAACAGCAATCAAGCGCGGAGAAGATAGAGGCTGCGATGGACGACAGCGCCGTGAGACGGTTCTTCGACTGCGTGCTGCAGCACATCGACAACGGGACCCTCTCCACCGCCGTGAGGCTGAACATGCTGCGCTCGGCCGAGGAACAACTTAACAAGTTGTCGGAGATGGAGCCGGCGGTGTGCGGCGCGGCGCAGCGGACGGCACTGTTCGCGCGCTGCGTGCGGGCGCTGCATGACGCGCTGCAGTGCGCCGCGCCGCCGCAGCACGCGCTGCGCTCCCTGCACGCAGACTGTCTCCGGTCGGTAGCCTCCATCATTACATAA
Protein
MAAVLKKRALAEYNKSFQDAPPSKKLHLAKKPLIGSSAAAFVGLLEKCKSSDEALQFLLRISDCLQFQESDVEEAIKKLSKHFQSEEESVVRVKILWLFCDIGLEVPCANLNNLIDETIHLIKNETSHKVIGQGIATLTKLAKKLSDDKILMMRLVGVAKDNLKDTSHQVKCRCLQLISELYPIHPDSDRTSEMTAEAEAIVKLLGDYSNSEDARVRCEAFQALLTLHQRGQILGSNLYEHVCVALSDDYEIVREVALKLIWVLGNKYPENVITLQDDETTIRMVDDAFVRICSAVNDLCMQVRALACTLLGTTRAVSDRFLLQTLDKQLMSNMKKKRTAHERGAELVRSGAWASGRRWADDAPGALVPSAAVQLLPGGAAGAFVHGLEDELMEVRTAAVEALCQLSLENPVFATTSLDFLVDMFNDEIEEVRLRAIDALTRISHHIVLREDLLETILGALEDYSMDVREGLHRLLGSCTVASKTCLEMCIDKILENLKRYPQDKRSTFRCVQRLGSSHATLVLPLTTRLLAVHPFFDMPEPDVEDPAYMCVLILVLNAAQHCTTMLPLFEEHTIKHYTYLRDTMPHLVPHLPIGEQQSSAEKIEAAMDDSAVRRFFDCVLQHIDNGTLSTAVRLNMLRSAEEQLNKLSEMEPAVCGAAQRTALFARCVRALHDALQCAAPPQHALRSLHADCLRSVASIIT
Summary
Description
Component of the Integrator complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing.
Subunit
Belongs to the multiprotein complex Integrator.
Similarity
Belongs to the Integrator subunit 4 family.
Keywords
Nucleus
Repeat
Feature
chain Integrator complex subunit 4
Uniprot
A0A2H1WH17
A0A3S2NAW5
A0A194R938
A0A194PJB0
H9J1H8
A0A2A4JG33
+ More
A0A2J7PTS3 A0A1B6L0V8 A0A1B6E309 A0A1Y1KGH3 D6WNP8 A0A1B6FTP8 A0A069DWU3 A0A023F1P2 A0A0V0GA73 T1HLK5 A0A224XIX8 A0A0A9XLV2 E0VWN5 N6T0P4 U4U0A7 A0A2R7WQR2 A0A182GIB1 A0A182JG55 Q17LI2 A0A182VHR8 A0A182M153 A0A1Q3F7B9 A0A182PRP9 A0A182X9J0 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182NQM0 A0A182QT24 A0A182K522 A0A182VVV1 A0A084VXS1 A0A1B0CCD7 A0A182RVR1 A0A182YBF8 A0A1B0D7Z6 A0A224YTP1 A0A131Z4A6 L7M7B4 B0X7L2 Q7PX93 A0A2M4AHN6 A0A182FRM5 W5J837 A0A2R5LJB7 A0A336L9B2 A0A336LYV8 A0A147BQL0 A0A023GLA3 A0A1E1XBR9 A0A0P6B3A9 A0A0P5ZH62 A0A0P6EJX1 A0A0P4Y679 A0A0N8CNJ6 A0A0P5W2Y5 A0A293LIB9 A0A0N8D876 A0A0P5W899 A0A0P4XS64 A0A0N8CKH6 A0A131XES2 A0A1E1XM30 A0A0P5U6C4 E9HE00 A0A0A1WXA9 V5HG76 V3ZV05 A0A034V2J9 A0A0K8U138 C3Z8B9 A0A0P5Z5T2 W8BMN1 A0A1J1IDZ5 W5MHD2 A0A151N5H4 A0A151N4X6 Q68F70 A0A1A9X5K4 A0A3Q0FVC5 A0A1U7SVQ9 F6UCW4 A0A3Q0FQW1 A0A0P6E5N1 A0A1B0A5E6 A0A2Y9RFT3 B3NVG4 A0A240SWV1 A0A452EWM9 W5P9W4 K7F963
A0A2J7PTS3 A0A1B6L0V8 A0A1B6E309 A0A1Y1KGH3 D6WNP8 A0A1B6FTP8 A0A069DWU3 A0A023F1P2 A0A0V0GA73 T1HLK5 A0A224XIX8 A0A0A9XLV2 E0VWN5 N6T0P4 U4U0A7 A0A2R7WQR2 A0A182GIB1 A0A182JG55 Q17LI2 A0A182VHR8 A0A182M153 A0A1Q3F7B9 A0A182PRP9 A0A182X9J0 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182NQM0 A0A182QT24 A0A182K522 A0A182VVV1 A0A084VXS1 A0A1B0CCD7 A0A182RVR1 A0A182YBF8 A0A1B0D7Z6 A0A224YTP1 A0A131Z4A6 L7M7B4 B0X7L2 Q7PX93 A0A2M4AHN6 A0A182FRM5 W5J837 A0A2R5LJB7 A0A336L9B2 A0A336LYV8 A0A147BQL0 A0A023GLA3 A0A1E1XBR9 A0A0P6B3A9 A0A0P5ZH62 A0A0P6EJX1 A0A0P4Y679 A0A0N8CNJ6 A0A0P5W2Y5 A0A293LIB9 A0A0N8D876 A0A0P5W899 A0A0P4XS64 A0A0N8CKH6 A0A131XES2 A0A1E1XM30 A0A0P5U6C4 E9HE00 A0A0A1WXA9 V5HG76 V3ZV05 A0A034V2J9 A0A0K8U138 C3Z8B9 A0A0P5Z5T2 W8BMN1 A0A1J1IDZ5 W5MHD2 A0A151N5H4 A0A151N4X6 Q68F70 A0A1A9X5K4 A0A3Q0FVC5 A0A1U7SVQ9 F6UCW4 A0A3Q0FQW1 A0A0P6E5N1 A0A1B0A5E6 A0A2Y9RFT3 B3NVG4 A0A240SWV1 A0A452EWM9 W5P9W4 K7F963
Pubmed
26354079
19121390
28004739
18362917
19820115
26334808
+ More
25474469 25401762 26823975 20566863 23537049 26483478 17510324 20966253 24438588 25244985 28797301 26830274 25576852 12364791 14747013 17210077 20920257 23761445 29652888 28503490 28049606 29209593 21292972 25830018 25765539 23254933 25348373 18563158 24495485 22293439 20431018 17994087 20809919 17381049
25474469 25401762 26823975 20566863 23537049 26483478 17510324 20966253 24438588 25244985 28797301 26830274 25576852 12364791 14747013 17210077 20920257 23761445 29652888 28503490 28049606 29209593 21292972 25830018 25765539 23254933 25348373 18563158 24495485 22293439 20431018 17994087 20809919 17381049
EMBL
ODYU01008620
SOQ52359.1
RSAL01000300
RVE42754.1
KQ460615
KPJ13770.1
+ More
KQ459602 KPI93511.1 BABH01023717 BABH01023718 BABH01023719 NWSH01001620 PCG70668.1 NEVH01021221 PNF19738.1 GEBQ01022716 JAT17261.1 GEDC01005019 JAS32279.1 GEZM01088909 JAV57837.1 KQ971342 EFA03749.1 GECZ01016201 JAS53568.1 GBGD01000361 JAC88528.1 GBBI01003798 JAC14914.1 GECL01001444 JAP04680.1 ACPB03014046 GFTR01007884 JAW08542.1 GBHO01021907 GDHC01006813 JAG21697.1 JAQ11816.1 DS235823 EEB17791.1 APGK01049450 APGK01049451 APGK01049452 APGK01049453 APGK01049454 APGK01049455 KB741154 ENN73654.1 KB630450 KB631015 KB631016 KB631106 ERL83607.1 ERL84006.1 ERL84007.1 ERL84111.1 KK855304 PTY21912.1 JXUM01065564 KQ562354 KXJ76091.1 CH477215 EAT47554.1 AXCM01000928 GFDL01011593 JAV23452.1 APCN01000830 AXCN02001514 ATLV01018205 KE525224 KFB42765.1 AJWK01006638 AJVK01034900 GFPF01009149 MAA20295.1 GEDV01002962 JAP85595.1 GACK01005277 JAA59757.1 DS232454 EDS42024.1 AAAB01008987 EAA01205.5 GGFK01006969 MBW40290.1 ADMH02002105 ETN59025.1 GGLE01005389 MBY09515.1 UFQS01002143 UFQT01002143 SSX13317.1 SSX32753.1 UFQT01000194 SSX21437.1 GEGO01002364 JAR93040.1 GBBM01000726 JAC34692.1 GFAC01002539 JAT96649.1 GDIP01021913 JAM81802.1 GDIP01044587 JAM59128.1 GDIQ01061693 LRGB01002451 JAN33044.1 KZS07644.1 GDIP01235762 JAI87639.1 GDIP01114279 JAL89435.1 GDIP01105828 JAL97886.1 GFWV01002869 MAA27599.1 GDIP01059239 JAM44476.1 GDIP01103085 JAM00630.1 GDIP01237366 JAI86035.1 GDIP01122839 JAL80875.1 GEFH01003739 JAP64842.1 GFAA01003383 JAU00052.1 GDIP01121235 JAL82479.1 GL732626 EFX70011.1 GBXI01010805 JAD03487.1 GANP01001823 JAB82645.1 KB203083 ESO86380.1 GAKP01022250 GAKP01022248 JAC36704.1 GDHF01032264 JAI20050.1 GG666593 EEN51243.1 GDIP01061229 JAM42486.1 GAMC01006528 JAC00028.1 CVRI01000047 CRK98495.1 AHAT01003066 AKHW03004037 KYO31849.1 KYO31848.1 BC079973 AAMC01059354 AAMC01059355 GDIP01170020 GDIQ01082079 JAJ53382.1 JAN12658.1 CH954180 EDV46356.1 CCAG010000014 CCAG010000015 LWLT01000026 AMGL01060853 AMGL01060854 AMGL01060855 AMGL01060856 AMGL01060857 AMGL01060858 AMGL01060859 AMGL01060860 AGCU01145919 AGCU01145920 AGCU01145921
KQ459602 KPI93511.1 BABH01023717 BABH01023718 BABH01023719 NWSH01001620 PCG70668.1 NEVH01021221 PNF19738.1 GEBQ01022716 JAT17261.1 GEDC01005019 JAS32279.1 GEZM01088909 JAV57837.1 KQ971342 EFA03749.1 GECZ01016201 JAS53568.1 GBGD01000361 JAC88528.1 GBBI01003798 JAC14914.1 GECL01001444 JAP04680.1 ACPB03014046 GFTR01007884 JAW08542.1 GBHO01021907 GDHC01006813 JAG21697.1 JAQ11816.1 DS235823 EEB17791.1 APGK01049450 APGK01049451 APGK01049452 APGK01049453 APGK01049454 APGK01049455 KB741154 ENN73654.1 KB630450 KB631015 KB631016 KB631106 ERL83607.1 ERL84006.1 ERL84007.1 ERL84111.1 KK855304 PTY21912.1 JXUM01065564 KQ562354 KXJ76091.1 CH477215 EAT47554.1 AXCM01000928 GFDL01011593 JAV23452.1 APCN01000830 AXCN02001514 ATLV01018205 KE525224 KFB42765.1 AJWK01006638 AJVK01034900 GFPF01009149 MAA20295.1 GEDV01002962 JAP85595.1 GACK01005277 JAA59757.1 DS232454 EDS42024.1 AAAB01008987 EAA01205.5 GGFK01006969 MBW40290.1 ADMH02002105 ETN59025.1 GGLE01005389 MBY09515.1 UFQS01002143 UFQT01002143 SSX13317.1 SSX32753.1 UFQT01000194 SSX21437.1 GEGO01002364 JAR93040.1 GBBM01000726 JAC34692.1 GFAC01002539 JAT96649.1 GDIP01021913 JAM81802.1 GDIP01044587 JAM59128.1 GDIQ01061693 LRGB01002451 JAN33044.1 KZS07644.1 GDIP01235762 JAI87639.1 GDIP01114279 JAL89435.1 GDIP01105828 JAL97886.1 GFWV01002869 MAA27599.1 GDIP01059239 JAM44476.1 GDIP01103085 JAM00630.1 GDIP01237366 JAI86035.1 GDIP01122839 JAL80875.1 GEFH01003739 JAP64842.1 GFAA01003383 JAU00052.1 GDIP01121235 JAL82479.1 GL732626 EFX70011.1 GBXI01010805 JAD03487.1 GANP01001823 JAB82645.1 KB203083 ESO86380.1 GAKP01022250 GAKP01022248 JAC36704.1 GDHF01032264 JAI20050.1 GG666593 EEN51243.1 GDIP01061229 JAM42486.1 GAMC01006528 JAC00028.1 CVRI01000047 CRK98495.1 AHAT01003066 AKHW03004037 KYO31849.1 KYO31848.1 BC079973 AAMC01059354 AAMC01059355 GDIP01170020 GDIQ01082079 JAJ53382.1 JAN12658.1 CH954180 EDV46356.1 CCAG010000014 CCAG010000015 LWLT01000026 AMGL01060853 AMGL01060854 AMGL01060855 AMGL01060856 AMGL01060857 AMGL01060858 AMGL01060859 AMGL01060860 AGCU01145919 AGCU01145920 AGCU01145921
Proteomes
UP000283053
UP000053240
UP000053268
UP000005204
UP000218220
UP000235965
+ More
UP000007266 UP000015103 UP000009046 UP000019118 UP000030742 UP000069940 UP000249989 UP000075880 UP000008820 UP000075903 UP000075883 UP000075885 UP000076407 UP000075882 UP000075902 UP000075840 UP000075884 UP000075886 UP000075881 UP000075920 UP000030765 UP000092461 UP000075900 UP000076408 UP000092462 UP000002320 UP000007062 UP000069272 UP000000673 UP000076858 UP000000305 UP000030746 UP000001554 UP000183832 UP000018468 UP000050525 UP000091820 UP000189705 UP000008143 UP000092445 UP000248480 UP000008711 UP000092444 UP000291000 UP000002356 UP000007267
UP000007266 UP000015103 UP000009046 UP000019118 UP000030742 UP000069940 UP000249989 UP000075880 UP000008820 UP000075903 UP000075883 UP000075885 UP000076407 UP000075882 UP000075902 UP000075840 UP000075884 UP000075886 UP000075881 UP000075920 UP000030765 UP000092461 UP000075900 UP000076408 UP000092462 UP000002320 UP000007062 UP000069272 UP000000673 UP000076858 UP000000305 UP000030746 UP000001554 UP000183832 UP000018468 UP000050525 UP000091820 UP000189705 UP000008143 UP000092445 UP000248480 UP000008711 UP000092444 UP000291000 UP000002356 UP000007267
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2H1WH17
A0A3S2NAW5
A0A194R938
A0A194PJB0
H9J1H8
A0A2A4JG33
+ More
A0A2J7PTS3 A0A1B6L0V8 A0A1B6E309 A0A1Y1KGH3 D6WNP8 A0A1B6FTP8 A0A069DWU3 A0A023F1P2 A0A0V0GA73 T1HLK5 A0A224XIX8 A0A0A9XLV2 E0VWN5 N6T0P4 U4U0A7 A0A2R7WQR2 A0A182GIB1 A0A182JG55 Q17LI2 A0A182VHR8 A0A182M153 A0A1Q3F7B9 A0A182PRP9 A0A182X9J0 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182NQM0 A0A182QT24 A0A182K522 A0A182VVV1 A0A084VXS1 A0A1B0CCD7 A0A182RVR1 A0A182YBF8 A0A1B0D7Z6 A0A224YTP1 A0A131Z4A6 L7M7B4 B0X7L2 Q7PX93 A0A2M4AHN6 A0A182FRM5 W5J837 A0A2R5LJB7 A0A336L9B2 A0A336LYV8 A0A147BQL0 A0A023GLA3 A0A1E1XBR9 A0A0P6B3A9 A0A0P5ZH62 A0A0P6EJX1 A0A0P4Y679 A0A0N8CNJ6 A0A0P5W2Y5 A0A293LIB9 A0A0N8D876 A0A0P5W899 A0A0P4XS64 A0A0N8CKH6 A0A131XES2 A0A1E1XM30 A0A0P5U6C4 E9HE00 A0A0A1WXA9 V5HG76 V3ZV05 A0A034V2J9 A0A0K8U138 C3Z8B9 A0A0P5Z5T2 W8BMN1 A0A1J1IDZ5 W5MHD2 A0A151N5H4 A0A151N4X6 Q68F70 A0A1A9X5K4 A0A3Q0FVC5 A0A1U7SVQ9 F6UCW4 A0A3Q0FQW1 A0A0P6E5N1 A0A1B0A5E6 A0A2Y9RFT3 B3NVG4 A0A240SWV1 A0A452EWM9 W5P9W4 K7F963
A0A2J7PTS3 A0A1B6L0V8 A0A1B6E309 A0A1Y1KGH3 D6WNP8 A0A1B6FTP8 A0A069DWU3 A0A023F1P2 A0A0V0GA73 T1HLK5 A0A224XIX8 A0A0A9XLV2 E0VWN5 N6T0P4 U4U0A7 A0A2R7WQR2 A0A182GIB1 A0A182JG55 Q17LI2 A0A182VHR8 A0A182M153 A0A1Q3F7B9 A0A182PRP9 A0A182X9J0 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182NQM0 A0A182QT24 A0A182K522 A0A182VVV1 A0A084VXS1 A0A1B0CCD7 A0A182RVR1 A0A182YBF8 A0A1B0D7Z6 A0A224YTP1 A0A131Z4A6 L7M7B4 B0X7L2 Q7PX93 A0A2M4AHN6 A0A182FRM5 W5J837 A0A2R5LJB7 A0A336L9B2 A0A336LYV8 A0A147BQL0 A0A023GLA3 A0A1E1XBR9 A0A0P6B3A9 A0A0P5ZH62 A0A0P6EJX1 A0A0P4Y679 A0A0N8CNJ6 A0A0P5W2Y5 A0A293LIB9 A0A0N8D876 A0A0P5W899 A0A0P4XS64 A0A0N8CKH6 A0A131XES2 A0A1E1XM30 A0A0P5U6C4 E9HE00 A0A0A1WXA9 V5HG76 V3ZV05 A0A034V2J9 A0A0K8U138 C3Z8B9 A0A0P5Z5T2 W8BMN1 A0A1J1IDZ5 W5MHD2 A0A151N5H4 A0A151N4X6 Q68F70 A0A1A9X5K4 A0A3Q0FVC5 A0A1U7SVQ9 F6UCW4 A0A3Q0FQW1 A0A0P6E5N1 A0A1B0A5E6 A0A2Y9RFT3 B3NVG4 A0A240SWV1 A0A452EWM9 W5P9W4 K7F963
Ontologies
GO
Topology
Subcellular location
Nucleus
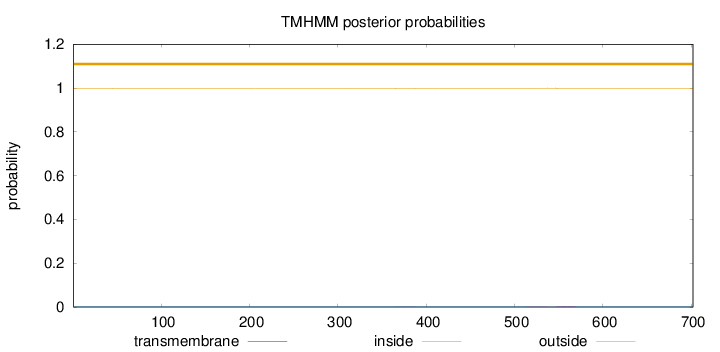
Length:
702
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06404
Exp number, first 60 AAs:
0.00152
Total prob of N-in:
0.00134
outside
1 - 702
Population Genetic Test Statistics
Pi
214.24123
Theta
183.182724
Tajima's D
0.63782
CLR
0.347667
CSRT
0.551322433878306
Interpretation
Uncertain
