Gene
KWMTBOMO09173
Pre Gene Modal
BGIBMGA003366
Annotation
PREDICTED:_integrator_complex_subunit_4_[Papilio_xuthus]
Full name
Integrator complex subunit 4
Location in the cell
Mitochondrial Reliability : 1.173
Sequence
CDS
ATGTCCGAACAGGACAATGCGTGCGTTTGGCAGCTGGCGCTGCGAGTGTGCGCCGCCCGCCTGTGCGCCGCCGTGCAGCAGAGCGCGGCTGGCAACACTAGCGGCGGGGTCGCTGCAGCTGGCAACACCGCCGCGCTGTCGGCCGTCGCAGCTCTCACGCACCACGCCGAAGTGTTGGACAGAATGCTGGCTTCCGCAGGTGTAGAGCCGGATCCGTTCACGATAGCAGTGTTCCGTCAGCTGTCGATGAACGCGGACAACAAGCCGGCCGCGCTGGCGAGGAGCATCCACCCGCTCCTACTGTCCGCGCCGCTGCCGGTCATACCCAAACCTAACCTTAAGATCGTGATGTGCAAAGCTAGCATAGTGGAGCCGGCCGCCGACAACGACTTGGTGGTGCGGTTCAGCGCGGGGCTCGTGGCCGGCGTGGCGCTGGAGGCCGAGGTGACGCGCGTGCGCTGTGCGGACGCGCTGCGCGTGCGCGTGGCCTACCCGGACCGCCGGGTGCACGCGCTGCTGCCGCCCCGGGACCACCTCCGGCCGCTCGACCATCACGACCAAAATGAGGATGGAACGCAAAACTTGCGTTTGCTGACTAAAGTTTTAATTTCACATGGAGTGTGGTCAGAACCATGTGGAGTGGACATTTCTATATGCCTAGCGGTCGAAGAAGGCGGTGGCAGAGAACCACCACAACTAGTAGAGCTTTGTAAACCAGTTAGAGTTACTGTAGCACCAAAGCCTATCAAACGAGGAATATAA
Protein
MSEQDNACVWQLALRVCAARLCAAVQQSAAGNTSGGVAAAGNTAALSAVAALTHHAEVLDRMLASAGVEPDPFTIAVFRQLSMNADNKPAALARSIHPLLLSAPLPVIPKPNLKIVMCKASIVEPAADNDLVVRFSAGLVAGVALEAEVTRVRCADALRVRVAYPDRRVHALLPPRDHLRPLDHHDQNEDGTQNLRLLTKVLISHGVWSEPCGVDISICLAVEEGGGREPPQLVELCKPVRVTVAPKPIKRGI
Summary
Description
Component of the Integrator complex, a complex involved in the transcription of small nuclear RNAs (snRNA) and their 3'-box-dependent processing (PubMed:21078872, PubMed:23097424). Involved in the 3'-end processing of the U7 snRNA, and also the spliceosomal snRNAs U1, U2, U4 and U5 (PubMed:21078872, PubMed:23097424). Essential for development (PubMed:21078872). May mediate recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (By similarity).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of IntS1, IntS2, IntS3, IntS4, omd/IntS5, IntS6, defl/IntS7, IntS8, IntS9, IntS10, IntS11, IntS12, asun/IntS13 and IntS14.
Similarity
Belongs to the Integrator subunit 4 family.
Keywords
Complete proteome
Nucleus
Reference proteome
Feature
chain Integrator complex subunit 4
Uniprot
H9J1H9
A0A2A4JG33
A0A2H1WH17
A0A3S2NAW5
A0A194PJB0
A0A194R938
+ More
A0A1Y1KGH3 D6WNP8 E0VWN5 N6T0P4 A0A067R8X0 A0A0K8SS89 A0A146KUM2 A0A0A9XLV2 A0A2J7PTT1 A0A2J7PTS3 A0A2R7WQR2 A0A1B6KFJ8 T1HLK5 A0A1B6EUS6 A0A069DWU3 A0A023F1P2 A0A1B6JDN2 A0A2P8Z301 A0A0V0GA73 B7QNR7 A0A293LIB9 A0A1S3DQ96 A0A182SNE1 A0A1Q3F7B9 A0A182NQM0 A0A182QT24 A0A087UJI0 A0A084VXS1 A0A182X9J0 A0A182PRP9 A0A1B6E309 A0A182JG55 A0A182VHR8 A0A182VVV1 Q7PX93 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182YBF8 A0A182RVR1 Q17LI2 A0A2L2YE14 A0A182K522 B0X7L2 A0A182GIB1 B4HA28 A0A147BQL0 V5HG76 W5J837 A0A2M4AHN6 A0A182FRM5 A0A023GJC6 A0A224YTP1 A0A182M153 A0A131Z4A6 A0A423THR1 L7M7B4 A0A131XES2 A0A1B0AXN7 A0A1E1XM30 W8BMN1 A0A2P6K3Q8 Q29FM0 A0A0P4WD44 A0A1I8Q286 T1PPE2 A0A336LYV8 B4L4X5 B4M1M8 A0A1E1XBR9 A0A1W4VV02 A0A1B0A5E6 A0A1A9V6R0 A0A2R5LJB7 B3N0B1 A0A0A1WXA9 B4JN25 A0A1S3DQZ2 A0A1A9X5K4 A0A240SWV1 A0A1I8MZZ2 B4NCA9 B4IJX9 A0A0P4W6N6 A0A034V2J9 A0A0K8U138 B4Q0A1 Q9W3E1 A0A1D2N3C4 A0A1I8Q290
A0A1Y1KGH3 D6WNP8 E0VWN5 N6T0P4 A0A067R8X0 A0A0K8SS89 A0A146KUM2 A0A0A9XLV2 A0A2J7PTT1 A0A2J7PTS3 A0A2R7WQR2 A0A1B6KFJ8 T1HLK5 A0A1B6EUS6 A0A069DWU3 A0A023F1P2 A0A1B6JDN2 A0A2P8Z301 A0A0V0GA73 B7QNR7 A0A293LIB9 A0A1S3DQ96 A0A182SNE1 A0A1Q3F7B9 A0A182NQM0 A0A182QT24 A0A087UJI0 A0A084VXS1 A0A182X9J0 A0A182PRP9 A0A1B6E309 A0A182JG55 A0A182VHR8 A0A182VVV1 Q7PX93 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182YBF8 A0A182RVR1 Q17LI2 A0A2L2YE14 A0A182K522 B0X7L2 A0A182GIB1 B4HA28 A0A147BQL0 V5HG76 W5J837 A0A2M4AHN6 A0A182FRM5 A0A023GJC6 A0A224YTP1 A0A182M153 A0A131Z4A6 A0A423THR1 L7M7B4 A0A131XES2 A0A1B0AXN7 A0A1E1XM30 W8BMN1 A0A2P6K3Q8 Q29FM0 A0A0P4WD44 A0A1I8Q286 T1PPE2 A0A336LYV8 B4L4X5 B4M1M8 A0A1E1XBR9 A0A1W4VV02 A0A1B0A5E6 A0A1A9V6R0 A0A2R5LJB7 B3N0B1 A0A0A1WXA9 B4JN25 A0A1S3DQZ2 A0A1A9X5K4 A0A240SWV1 A0A1I8MZZ2 B4NCA9 B4IJX9 A0A0P4W6N6 A0A034V2J9 A0A0K8U138 B4Q0A1 Q9W3E1 A0A1D2N3C4 A0A1I8Q290
Pubmed
19121390
26354079
28004739
18362917
19820115
20566863
+ More
23537049 24845553 26823975 25401762 26334808 25474469 29403074 24438588 12364791 14747013 17210077 20966253 25244985 17510324 26561354 26483478 17994087 29652888 25765539 20920257 23761445 28797301 26830274 25576852 28049606 29209593 24495485 15632085 28503490 25830018 25315136 25348373 17550304 10731132 12537572 21078872 23097424 27289101
23537049 24845553 26823975 25401762 26334808 25474469 29403074 24438588 12364791 14747013 17210077 20966253 25244985 17510324 26561354 26483478 17994087 29652888 25765539 20920257 23761445 28797301 26830274 25576852 28049606 29209593 24495485 15632085 28503490 25830018 25315136 25348373 17550304 10731132 12537572 21078872 23097424 27289101
EMBL
BABH01023716
BABH01023717
NWSH01001620
PCG70668.1
ODYU01008620
SOQ52359.1
+ More
RSAL01000300 RVE42754.1 KQ459602 KPI93511.1 KQ460615 KPJ13770.1 GEZM01088909 JAV57837.1 KQ971342 EFA03749.1 DS235823 EEB17791.1 APGK01049450 APGK01049451 APGK01049452 APGK01049453 APGK01049454 APGK01049455 KB741154 ENN73654.1 KK852619 KDR20101.1 GBRD01009800 JAG56024.1 GDHC01018438 GDHC01011408 JAQ00191.1 JAQ07221.1 GBHO01021907 GDHC01006813 JAG21697.1 JAQ11816.1 NEVH01021221 PNF19739.1 PNF19738.1 KK855304 PTY21912.1 GEBQ01029747 JAT10230.1 ACPB03014046 GECZ01028131 JAS41638.1 GBGD01000361 JAC88528.1 GBBI01003798 JAC14914.1 GECU01010376 JAS97330.1 PYGN01000219 PSN50884.1 GECL01001444 JAP04680.1 ABJB010057111 ABJB010192685 ABJB010283782 ABJB010306238 ABJB010382177 ABJB010414991 ABJB010632223 ABJB010708409 ABJB010738996 ABJB011062949 DS979863 EEC20489.1 GFWV01002869 MAA27599.1 GFDL01011593 JAV23452.1 AXCN02001514 KK120096 KFM77519.1 ATLV01018205 KE525224 KFB42765.1 GEDC01005019 JAS32279.1 AAAB01008987 EAA01205.5 APCN01000830 CH477215 EAT47554.1 IAAA01023360 LAA06374.1 DS232454 EDS42024.1 JXUM01065564 KQ562354 KXJ76091.1 CH479235 EDW36696.1 GEGO01002364 JAR93040.1 GANP01001823 JAB82645.1 ADMH02002105 ETN59025.1 GGFK01006969 MBW40290.1 GBBM01001366 JAC34052.1 GFPF01009149 MAA20295.1 AXCM01000928 GEDV01002962 JAP85595.1 QCYY01001702 ROT75993.1 GACK01005277 JAA59757.1 GEFH01003739 JAP64842.1 JXJN01005326 GFAA01003383 JAU00052.1 GAMC01006528 JAC00028.1 MWRG01032267 PRD20942.1 CH379066 EAL31380.1 GDRN01068023 JAI64262.1 KA649980 AFP64609.1 UFQT01000194 SSX21437.1 CH933810 EDW07603.1 CH940651 EDW65582.1 GFAC01002539 JAT96649.1 GGLE01005389 MBY09515.1 CH902640 EDV38315.1 GBXI01010805 JAD03487.1 CH916371 EDV92118.1 CCAG010000014 CCAG010000015 CH964239 EDW82468.1 CH480850 EDW51335.1 GDRN01068022 JAI64263.1 GAKP01022250 GAKP01022248 JAC36704.1 GDHF01032264 JAI20050.1 CM000162 EDX02238.1 AE014298 AY051460 LJIJ01000256 ODM99768.1
RSAL01000300 RVE42754.1 KQ459602 KPI93511.1 KQ460615 KPJ13770.1 GEZM01088909 JAV57837.1 KQ971342 EFA03749.1 DS235823 EEB17791.1 APGK01049450 APGK01049451 APGK01049452 APGK01049453 APGK01049454 APGK01049455 KB741154 ENN73654.1 KK852619 KDR20101.1 GBRD01009800 JAG56024.1 GDHC01018438 GDHC01011408 JAQ00191.1 JAQ07221.1 GBHO01021907 GDHC01006813 JAG21697.1 JAQ11816.1 NEVH01021221 PNF19739.1 PNF19738.1 KK855304 PTY21912.1 GEBQ01029747 JAT10230.1 ACPB03014046 GECZ01028131 JAS41638.1 GBGD01000361 JAC88528.1 GBBI01003798 JAC14914.1 GECU01010376 JAS97330.1 PYGN01000219 PSN50884.1 GECL01001444 JAP04680.1 ABJB010057111 ABJB010192685 ABJB010283782 ABJB010306238 ABJB010382177 ABJB010414991 ABJB010632223 ABJB010708409 ABJB010738996 ABJB011062949 DS979863 EEC20489.1 GFWV01002869 MAA27599.1 GFDL01011593 JAV23452.1 AXCN02001514 KK120096 KFM77519.1 ATLV01018205 KE525224 KFB42765.1 GEDC01005019 JAS32279.1 AAAB01008987 EAA01205.5 APCN01000830 CH477215 EAT47554.1 IAAA01023360 LAA06374.1 DS232454 EDS42024.1 JXUM01065564 KQ562354 KXJ76091.1 CH479235 EDW36696.1 GEGO01002364 JAR93040.1 GANP01001823 JAB82645.1 ADMH02002105 ETN59025.1 GGFK01006969 MBW40290.1 GBBM01001366 JAC34052.1 GFPF01009149 MAA20295.1 AXCM01000928 GEDV01002962 JAP85595.1 QCYY01001702 ROT75993.1 GACK01005277 JAA59757.1 GEFH01003739 JAP64842.1 JXJN01005326 GFAA01003383 JAU00052.1 GAMC01006528 JAC00028.1 MWRG01032267 PRD20942.1 CH379066 EAL31380.1 GDRN01068023 JAI64262.1 KA649980 AFP64609.1 UFQT01000194 SSX21437.1 CH933810 EDW07603.1 CH940651 EDW65582.1 GFAC01002539 JAT96649.1 GGLE01005389 MBY09515.1 CH902640 EDV38315.1 GBXI01010805 JAD03487.1 CH916371 EDV92118.1 CCAG010000014 CCAG010000015 CH964239 EDW82468.1 CH480850 EDW51335.1 GDRN01068022 JAI64263.1 GAKP01022250 GAKP01022248 JAC36704.1 GDHF01032264 JAI20050.1 CM000162 EDX02238.1 AE014298 AY051460 LJIJ01000256 ODM99768.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007266
+ More
UP000009046 UP000019118 UP000027135 UP000235965 UP000015103 UP000245037 UP000001555 UP000079169 UP000075901 UP000075884 UP000075886 UP000054359 UP000030765 UP000076407 UP000075885 UP000075880 UP000075903 UP000075920 UP000007062 UP000075882 UP000075902 UP000075840 UP000076408 UP000075900 UP000008820 UP000075881 UP000002320 UP000069940 UP000249989 UP000008744 UP000000673 UP000069272 UP000075883 UP000283509 UP000092460 UP000001819 UP000095300 UP000009192 UP000008792 UP000192221 UP000092445 UP000078200 UP000007801 UP000001070 UP000091820 UP000092444 UP000095301 UP000007798 UP000001292 UP000002282 UP000000803 UP000094527
UP000009046 UP000019118 UP000027135 UP000235965 UP000015103 UP000245037 UP000001555 UP000079169 UP000075901 UP000075884 UP000075886 UP000054359 UP000030765 UP000076407 UP000075885 UP000075880 UP000075903 UP000075920 UP000007062 UP000075882 UP000075902 UP000075840 UP000076408 UP000075900 UP000008820 UP000075881 UP000002320 UP000069940 UP000249989 UP000008744 UP000000673 UP000069272 UP000075883 UP000283509 UP000092460 UP000001819 UP000095300 UP000009192 UP000008792 UP000192221 UP000092445 UP000078200 UP000007801 UP000001070 UP000091820 UP000092444 UP000095301 UP000007798 UP000001292 UP000002282 UP000000803 UP000094527
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9J1H9
A0A2A4JG33
A0A2H1WH17
A0A3S2NAW5
A0A194PJB0
A0A194R938
+ More
A0A1Y1KGH3 D6WNP8 E0VWN5 N6T0P4 A0A067R8X0 A0A0K8SS89 A0A146KUM2 A0A0A9XLV2 A0A2J7PTT1 A0A2J7PTS3 A0A2R7WQR2 A0A1B6KFJ8 T1HLK5 A0A1B6EUS6 A0A069DWU3 A0A023F1P2 A0A1B6JDN2 A0A2P8Z301 A0A0V0GA73 B7QNR7 A0A293LIB9 A0A1S3DQ96 A0A182SNE1 A0A1Q3F7B9 A0A182NQM0 A0A182QT24 A0A087UJI0 A0A084VXS1 A0A182X9J0 A0A182PRP9 A0A1B6E309 A0A182JG55 A0A182VHR8 A0A182VVV1 Q7PX93 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182YBF8 A0A182RVR1 Q17LI2 A0A2L2YE14 A0A182K522 B0X7L2 A0A182GIB1 B4HA28 A0A147BQL0 V5HG76 W5J837 A0A2M4AHN6 A0A182FRM5 A0A023GJC6 A0A224YTP1 A0A182M153 A0A131Z4A6 A0A423THR1 L7M7B4 A0A131XES2 A0A1B0AXN7 A0A1E1XM30 W8BMN1 A0A2P6K3Q8 Q29FM0 A0A0P4WD44 A0A1I8Q286 T1PPE2 A0A336LYV8 B4L4X5 B4M1M8 A0A1E1XBR9 A0A1W4VV02 A0A1B0A5E6 A0A1A9V6R0 A0A2R5LJB7 B3N0B1 A0A0A1WXA9 B4JN25 A0A1S3DQZ2 A0A1A9X5K4 A0A240SWV1 A0A1I8MZZ2 B4NCA9 B4IJX9 A0A0P4W6N6 A0A034V2J9 A0A0K8U138 B4Q0A1 Q9W3E1 A0A1D2N3C4 A0A1I8Q290
A0A1Y1KGH3 D6WNP8 E0VWN5 N6T0P4 A0A067R8X0 A0A0K8SS89 A0A146KUM2 A0A0A9XLV2 A0A2J7PTT1 A0A2J7PTS3 A0A2R7WQR2 A0A1B6KFJ8 T1HLK5 A0A1B6EUS6 A0A069DWU3 A0A023F1P2 A0A1B6JDN2 A0A2P8Z301 A0A0V0GA73 B7QNR7 A0A293LIB9 A0A1S3DQ96 A0A182SNE1 A0A1Q3F7B9 A0A182NQM0 A0A182QT24 A0A087UJI0 A0A084VXS1 A0A182X9J0 A0A182PRP9 A0A1B6E309 A0A182JG55 A0A182VHR8 A0A182VVV1 Q7PX93 A0A182L8R1 A0A182UJ60 A0A182HKH3 A0A182YBF8 A0A182RVR1 Q17LI2 A0A2L2YE14 A0A182K522 B0X7L2 A0A182GIB1 B4HA28 A0A147BQL0 V5HG76 W5J837 A0A2M4AHN6 A0A182FRM5 A0A023GJC6 A0A224YTP1 A0A182M153 A0A131Z4A6 A0A423THR1 L7M7B4 A0A131XES2 A0A1B0AXN7 A0A1E1XM30 W8BMN1 A0A2P6K3Q8 Q29FM0 A0A0P4WD44 A0A1I8Q286 T1PPE2 A0A336LYV8 B4L4X5 B4M1M8 A0A1E1XBR9 A0A1W4VV02 A0A1B0A5E6 A0A1A9V6R0 A0A2R5LJB7 B3N0B1 A0A0A1WXA9 B4JN25 A0A1S3DQZ2 A0A1A9X5K4 A0A240SWV1 A0A1I8MZZ2 B4NCA9 B4IJX9 A0A0P4W6N6 A0A034V2J9 A0A0K8U138 B4Q0A1 Q9W3E1 A0A1D2N3C4 A0A1I8Q290
Ontologies
Topology
Subcellular location
Nucleus
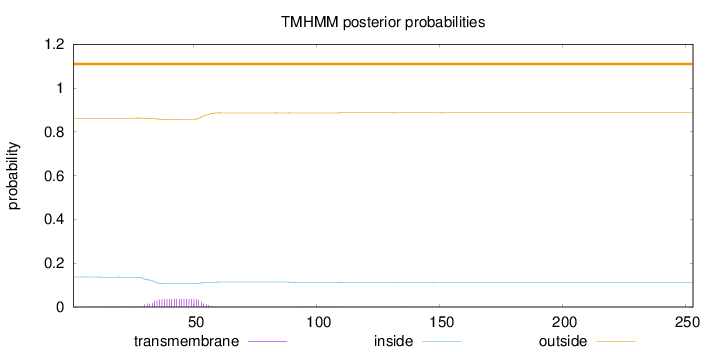
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.86636
Exp number, first 60 AAs:
0.81901
Total prob of N-in:
0.13754
outside
1 - 253
Population Genetic Test Statistics
Pi
130.683935
Theta
69.445176
Tajima's D
1.077504
CLR
0
CSRT
0.68111594420279
Interpretation
Uncertain
