Gene
KWMTBOMO09171 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003367
Annotation
stromal_cell-derived_factor_2_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.11
Sequence
CDS
ATGGAAAATACAAAAATCCTTTCAATTGCGACTTTAGTCACTGTGGTGTTTCTCATTTCTATTATAAGTGAAAAAACGGAAGCTGCGAAGAATGAATTTGTGACGTGTGGATCAATATTAAAACTCATCAACACAGACTTGAAGTTAAGACTTCATTCGCATGATGTAAAGTATGGGTCTGGATCAGGACAGCAGTCTGTGACGGCGGTTGAGGTCTCTGATGATAACAATAGTCACTGGCTGGTTCGCCCTATGACAGGAGAAACCTGCAAGCGTGGTGCTCCGATAAAATGTAACACCAACATTAGACTTCAGCATGTAGCTACAAAGAAGAACTTGCATTCACATTTTTTTACCTCACCCCTCTCTGGCAATCAAGAGGTATCATGTTATGGAGACGATGAGGGTGAAGGGGACAGTGGAGACAATTGGACTGTGGTCTGCAACAATGACTACTGGAGGAGAGATACACCAGTAAAATTTAGACATGTTGATACTGGATCGTATCTTGCAGGCTCCGGGCGAACATTTGGTCGTCCCATCAATGGTCAAGGCGAGATTGTCGGAGTGAGTTCACAATATGGTGCTTACACTGACTGGCAAGCAAGTGAAGGTCTCTTTGTTCATCCTGGAGAACTATTACCACATCAGCATGCTGTACATACAGAGTTATAA
Protein
MENTKILSIATLVTVVFLISIISEKTEAAKNEFVTCGSILKLINTDLKLRLHSHDVKYGSGSGQQSVTAVEVSDDNNSHWLVRPMTGETCKRGAPIKCNTNIRLQHVATKKNLHSHFFTSPLSGNQEVSCYGDDEGEGDSGDNWTVVCNNDYWRRDTPVKFRHVDTGSYLAGSGRTFGRPINGQGEIVGVSSQYGAYTDWQASEGLFVHPGELLPHQHAVHTEL
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
Q2F5L6
A0A194PL98
A0A194R7G3
I4DKU9
A0A2A4JFC4
A0A2H1VUB8
+ More
F1C4B9 A0A1E1WVI1 A0A437AX71 A0A212F763 A0A0L7L1D3 S4PDU7 A0A1L8DVW8 A0A1Q3F9F0 B0XAX9 Q1HQT2 A0A1B0CKB7 A0A182G6G4 A0A023EJX9 D7EK01 A0A1J1IKE7 A0A023EJZ0 A0A182QCJ1 A0A084W306 A0A1Y1MXS6 Q17HC4 A0A1B0D2S3 A0A182ILY5 A0A182NLI9 A0A182VRS6 A0A1W4XKK7 A0A182LA67 Q7PY52 A0A182HLR6 A0A182VG04 A0A1L8EH29 W5JM99 A0A182WSW2 A0A2M4AAG7 A0A182XV01 A0A182UH10 A0A2M4C015 A0A2M4BYW4 A0A1I8NPT5 A0A182RPC8 A0A2M4BZ07 N6UH43 A0A182FNK5 T1DRB3 A0A2M3Z7M0 A0A1A9VG41 A0A1B0A1N2 D3TKL8 A0A1A9Y8D8 K7J8I9 A0A1B6J7F6 A0A182T8L8 A0A232FCB1 A0A182K8P0 A0A182MNK8 A0A182P2M3 A0A1B6FVD8 A0A1A9WX57 A0A023EJY1 A0A195E4T8 B3M181 A0A151X1N3 A0A336KR54 A0A034WLT9 A0A158NFK1 A0A0A1WMJ0 A0A0K8WHE2 B4QWA5 A0A3L8D480 A0A3B0JW14 B4JSN6 A0A026VVJ3 A0A1B0BMS4 B4M4W7 A0A195EUA4 A0A336MWV8 B3P2B7 A0A0M4ENN0 B4KC08 W8C9F1 A0A1I8MMV1 A0A0L0CDT9 A0A067QWG5 Q9VNA3 U5EJF4 B4I3Y7 A0A195CLG6 A0A1W4UWL4 Q296B4 B4GFD8 A0A0N0BIY4 A0A2A3ECT2 V9IBI0 C4WRN8 F4W484
F1C4B9 A0A1E1WVI1 A0A437AX71 A0A212F763 A0A0L7L1D3 S4PDU7 A0A1L8DVW8 A0A1Q3F9F0 B0XAX9 Q1HQT2 A0A1B0CKB7 A0A182G6G4 A0A023EJX9 D7EK01 A0A1J1IKE7 A0A023EJZ0 A0A182QCJ1 A0A084W306 A0A1Y1MXS6 Q17HC4 A0A1B0D2S3 A0A182ILY5 A0A182NLI9 A0A182VRS6 A0A1W4XKK7 A0A182LA67 Q7PY52 A0A182HLR6 A0A182VG04 A0A1L8EH29 W5JM99 A0A182WSW2 A0A2M4AAG7 A0A182XV01 A0A182UH10 A0A2M4C015 A0A2M4BYW4 A0A1I8NPT5 A0A182RPC8 A0A2M4BZ07 N6UH43 A0A182FNK5 T1DRB3 A0A2M3Z7M0 A0A1A9VG41 A0A1B0A1N2 D3TKL8 A0A1A9Y8D8 K7J8I9 A0A1B6J7F6 A0A182T8L8 A0A232FCB1 A0A182K8P0 A0A182MNK8 A0A182P2M3 A0A1B6FVD8 A0A1A9WX57 A0A023EJY1 A0A195E4T8 B3M181 A0A151X1N3 A0A336KR54 A0A034WLT9 A0A158NFK1 A0A0A1WMJ0 A0A0K8WHE2 B4QWA5 A0A3L8D480 A0A3B0JW14 B4JSN6 A0A026VVJ3 A0A1B0BMS4 B4M4W7 A0A195EUA4 A0A336MWV8 B3P2B7 A0A0M4ENN0 B4KC08 W8C9F1 A0A1I8MMV1 A0A0L0CDT9 A0A067QWG5 Q9VNA3 U5EJF4 B4I3Y7 A0A195CLG6 A0A1W4UWL4 Q296B4 B4GFD8 A0A0N0BIY4 A0A2A3ECT2 V9IBI0 C4WRN8 F4W484
Pubmed
19121390
26354079
22651552
22118469
26227816
23622113
+ More
17204158 26483478 24945155 18362917 19820115 24438588 28004739 17510324 20966253 12364791 14747013 17210077 20920257 23761445 25244985 23537049 20353571 20075255 28648823 17994087 25348373 21347285 25830018 30249741 24508170 24495485 25315136 26108605 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21719571
17204158 26483478 24945155 18362917 19820115 24438588 28004739 17510324 20966253 12364791 14747013 17210077 20920257 23761445 25244985 23537049 20353571 20075255 28648823 17994087 25348373 21347285 25830018 30249741 24508170 24495485 25315136 26108605 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 21719571
EMBL
BABH01023713
DQ311407
ABD36351.1
KQ459602
KPI93509.1
KQ460615
+ More
KPJ13768.1 AK401917 BAM18539.1 NWSH01001620 PCG70671.1 ODYU01004208 SOQ43834.1 HQ199329 ADX30520.1 GDQN01000015 JAT91039.1 RSAL01000300 RVE42752.1 AGBW02009919 OWR49573.1 JTDY01003700 KOB69121.1 GAIX01003496 JAA89064.1 GFDF01003512 JAV10572.1 GFDL01010902 JAV24143.1 DS232602 EDS43949.1 DQ440362 ABF18395.1 AJWK01015969 JXUM01045030 JXUM01045031 KQ561423 KXJ78631.1 GAPW01004187 JAC09411.1 DS497724 EFA12948.1 CVRI01000051 CRK99550.1 GAPW01004166 JAC09432.1 AXCN02000143 ATLV01019767 KE525279 KFB44600.1 GEZM01021945 JAV88806.1 CH477250 EAT46082.1 AJVK01023107 AAAB01008987 EAA00948.3 APCN01000911 GFDG01000821 JAV17978.1 ADMH02000703 ETN65276.1 GGFK01004434 MBW37755.1 GGFJ01009187 MBW58328.1 GGFJ01009144 MBW58285.1 GGFJ01009165 MBW58306.1 APGK01026291 KB740624 ENN79956.1 GAMD01000700 JAB00891.1 GGFM01003750 MBW24501.1 CCAG010009176 EZ421950 EZ424301 ADD18246.1 ADD20577.1 GECU01012614 JAS95092.1 NNAY01000478 OXU28103.1 AXCM01005967 GECZ01015650 JAS54119.1 GAPW01004186 JAC09412.1 KQ979609 KYN20178.1 CH902617 EDV44351.1 KQ982585 KYQ54309.1 UFQS01000733 UFQT01000733 SSX06471.1 SSX26820.1 GAKP01002406 JAC56546.1 ADTU01001535 GBXI01014431 JAC99860.1 GDHF01001738 JAI50576.1 CM000364 EDX11704.1 QOIP01000013 RLU15265.1 OUUW01000008 SPP84632.1 CH916373 EDV94776.1 KK107796 EZA47665.1 JXJN01017038 CH940652 EDW59678.1 KQ981979 KYN31459.1 UFQT01002491 SSX33569.1 CH954181 EDV47867.1 CP012526 ALC47252.1 CH933806 EDW15860.1 GAMC01002209 JAC04347.1 JRES01000531 KNC30406.1 KK853286 KDR08963.1 AE014297 AY118807 AAF52043.1 AAM50667.1 GANO01002254 JAB57617.1 CH480821 EDW54930.1 KQ977642 KYN00929.1 CM000070 EAL28544.1 CH479182 EDW34323.1 KQ435726 KOX78222.1 KZ288282 PBC29583.1 JR039030 AEY58493.1 ABLF02038286 AK339848 BAH70558.1 GL887491 EGI71080.1
KPJ13768.1 AK401917 BAM18539.1 NWSH01001620 PCG70671.1 ODYU01004208 SOQ43834.1 HQ199329 ADX30520.1 GDQN01000015 JAT91039.1 RSAL01000300 RVE42752.1 AGBW02009919 OWR49573.1 JTDY01003700 KOB69121.1 GAIX01003496 JAA89064.1 GFDF01003512 JAV10572.1 GFDL01010902 JAV24143.1 DS232602 EDS43949.1 DQ440362 ABF18395.1 AJWK01015969 JXUM01045030 JXUM01045031 KQ561423 KXJ78631.1 GAPW01004187 JAC09411.1 DS497724 EFA12948.1 CVRI01000051 CRK99550.1 GAPW01004166 JAC09432.1 AXCN02000143 ATLV01019767 KE525279 KFB44600.1 GEZM01021945 JAV88806.1 CH477250 EAT46082.1 AJVK01023107 AAAB01008987 EAA00948.3 APCN01000911 GFDG01000821 JAV17978.1 ADMH02000703 ETN65276.1 GGFK01004434 MBW37755.1 GGFJ01009187 MBW58328.1 GGFJ01009144 MBW58285.1 GGFJ01009165 MBW58306.1 APGK01026291 KB740624 ENN79956.1 GAMD01000700 JAB00891.1 GGFM01003750 MBW24501.1 CCAG010009176 EZ421950 EZ424301 ADD18246.1 ADD20577.1 GECU01012614 JAS95092.1 NNAY01000478 OXU28103.1 AXCM01005967 GECZ01015650 JAS54119.1 GAPW01004186 JAC09412.1 KQ979609 KYN20178.1 CH902617 EDV44351.1 KQ982585 KYQ54309.1 UFQS01000733 UFQT01000733 SSX06471.1 SSX26820.1 GAKP01002406 JAC56546.1 ADTU01001535 GBXI01014431 JAC99860.1 GDHF01001738 JAI50576.1 CM000364 EDX11704.1 QOIP01000013 RLU15265.1 OUUW01000008 SPP84632.1 CH916373 EDV94776.1 KK107796 EZA47665.1 JXJN01017038 CH940652 EDW59678.1 KQ981979 KYN31459.1 UFQT01002491 SSX33569.1 CH954181 EDV47867.1 CP012526 ALC47252.1 CH933806 EDW15860.1 GAMC01002209 JAC04347.1 JRES01000531 KNC30406.1 KK853286 KDR08963.1 AE014297 AY118807 AAF52043.1 AAM50667.1 GANO01002254 JAB57617.1 CH480821 EDW54930.1 KQ977642 KYN00929.1 CM000070 EAL28544.1 CH479182 EDW34323.1 KQ435726 KOX78222.1 KZ288282 PBC29583.1 JR039030 AEY58493.1 ABLF02038286 AK339848 BAH70558.1 GL887491 EGI71080.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
+ More
UP000037510 UP000002320 UP000092461 UP000069940 UP000249989 UP000007266 UP000183832 UP000075886 UP000030765 UP000008820 UP000092462 UP000075880 UP000075884 UP000075920 UP000192223 UP000075882 UP000007062 UP000075840 UP000075903 UP000000673 UP000076407 UP000076408 UP000075902 UP000095300 UP000075900 UP000019118 UP000069272 UP000078200 UP000092445 UP000092444 UP000092443 UP000002358 UP000075901 UP000215335 UP000075881 UP000075883 UP000075885 UP000091820 UP000078492 UP000007801 UP000075809 UP000005205 UP000000304 UP000279307 UP000268350 UP000001070 UP000053097 UP000092460 UP000008792 UP000078541 UP000008711 UP000092553 UP000009192 UP000095301 UP000037069 UP000027135 UP000000803 UP000001292 UP000078542 UP000192221 UP000001819 UP000008744 UP000053105 UP000242457 UP000007819 UP000007755
UP000037510 UP000002320 UP000092461 UP000069940 UP000249989 UP000007266 UP000183832 UP000075886 UP000030765 UP000008820 UP000092462 UP000075880 UP000075884 UP000075920 UP000192223 UP000075882 UP000007062 UP000075840 UP000075903 UP000000673 UP000076407 UP000076408 UP000075902 UP000095300 UP000075900 UP000019118 UP000069272 UP000078200 UP000092445 UP000092444 UP000092443 UP000002358 UP000075901 UP000215335 UP000075881 UP000075883 UP000075885 UP000091820 UP000078492 UP000007801 UP000075809 UP000005205 UP000000304 UP000279307 UP000268350 UP000001070 UP000053097 UP000092460 UP000008792 UP000078541 UP000008711 UP000092553 UP000009192 UP000095301 UP000037069 UP000027135 UP000000803 UP000001292 UP000078542 UP000192221 UP000001819 UP000008744 UP000053105 UP000242457 UP000007819 UP000007755
Interpro
Gene 3D
ProteinModelPortal
Q2F5L6
A0A194PL98
A0A194R7G3
I4DKU9
A0A2A4JFC4
A0A2H1VUB8
+ More
F1C4B9 A0A1E1WVI1 A0A437AX71 A0A212F763 A0A0L7L1D3 S4PDU7 A0A1L8DVW8 A0A1Q3F9F0 B0XAX9 Q1HQT2 A0A1B0CKB7 A0A182G6G4 A0A023EJX9 D7EK01 A0A1J1IKE7 A0A023EJZ0 A0A182QCJ1 A0A084W306 A0A1Y1MXS6 Q17HC4 A0A1B0D2S3 A0A182ILY5 A0A182NLI9 A0A182VRS6 A0A1W4XKK7 A0A182LA67 Q7PY52 A0A182HLR6 A0A182VG04 A0A1L8EH29 W5JM99 A0A182WSW2 A0A2M4AAG7 A0A182XV01 A0A182UH10 A0A2M4C015 A0A2M4BYW4 A0A1I8NPT5 A0A182RPC8 A0A2M4BZ07 N6UH43 A0A182FNK5 T1DRB3 A0A2M3Z7M0 A0A1A9VG41 A0A1B0A1N2 D3TKL8 A0A1A9Y8D8 K7J8I9 A0A1B6J7F6 A0A182T8L8 A0A232FCB1 A0A182K8P0 A0A182MNK8 A0A182P2M3 A0A1B6FVD8 A0A1A9WX57 A0A023EJY1 A0A195E4T8 B3M181 A0A151X1N3 A0A336KR54 A0A034WLT9 A0A158NFK1 A0A0A1WMJ0 A0A0K8WHE2 B4QWA5 A0A3L8D480 A0A3B0JW14 B4JSN6 A0A026VVJ3 A0A1B0BMS4 B4M4W7 A0A195EUA4 A0A336MWV8 B3P2B7 A0A0M4ENN0 B4KC08 W8C9F1 A0A1I8MMV1 A0A0L0CDT9 A0A067QWG5 Q9VNA3 U5EJF4 B4I3Y7 A0A195CLG6 A0A1W4UWL4 Q296B4 B4GFD8 A0A0N0BIY4 A0A2A3ECT2 V9IBI0 C4WRN8 F4W484
F1C4B9 A0A1E1WVI1 A0A437AX71 A0A212F763 A0A0L7L1D3 S4PDU7 A0A1L8DVW8 A0A1Q3F9F0 B0XAX9 Q1HQT2 A0A1B0CKB7 A0A182G6G4 A0A023EJX9 D7EK01 A0A1J1IKE7 A0A023EJZ0 A0A182QCJ1 A0A084W306 A0A1Y1MXS6 Q17HC4 A0A1B0D2S3 A0A182ILY5 A0A182NLI9 A0A182VRS6 A0A1W4XKK7 A0A182LA67 Q7PY52 A0A182HLR6 A0A182VG04 A0A1L8EH29 W5JM99 A0A182WSW2 A0A2M4AAG7 A0A182XV01 A0A182UH10 A0A2M4C015 A0A2M4BYW4 A0A1I8NPT5 A0A182RPC8 A0A2M4BZ07 N6UH43 A0A182FNK5 T1DRB3 A0A2M3Z7M0 A0A1A9VG41 A0A1B0A1N2 D3TKL8 A0A1A9Y8D8 K7J8I9 A0A1B6J7F6 A0A182T8L8 A0A232FCB1 A0A182K8P0 A0A182MNK8 A0A182P2M3 A0A1B6FVD8 A0A1A9WX57 A0A023EJY1 A0A195E4T8 B3M181 A0A151X1N3 A0A336KR54 A0A034WLT9 A0A158NFK1 A0A0A1WMJ0 A0A0K8WHE2 B4QWA5 A0A3L8D480 A0A3B0JW14 B4JSN6 A0A026VVJ3 A0A1B0BMS4 B4M4W7 A0A195EUA4 A0A336MWV8 B3P2B7 A0A0M4ENN0 B4KC08 W8C9F1 A0A1I8MMV1 A0A0L0CDT9 A0A067QWG5 Q9VNA3 U5EJF4 B4I3Y7 A0A195CLG6 A0A1W4UWL4 Q296B4 B4GFD8 A0A0N0BIY4 A0A2A3ECT2 V9IBI0 C4WRN8 F4W484
PDB
1T9F
E-value=6.98539e-45,
Score=452
Ontologies
GO
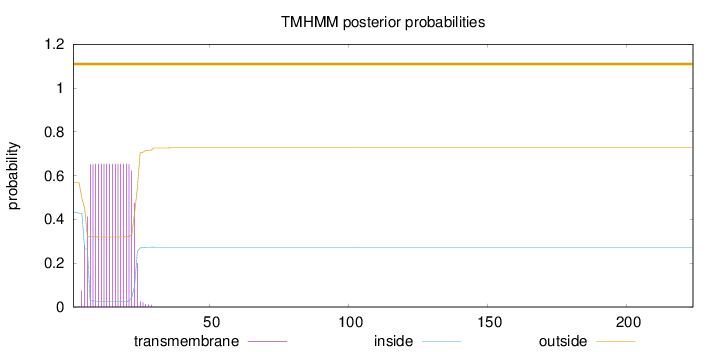
Topology
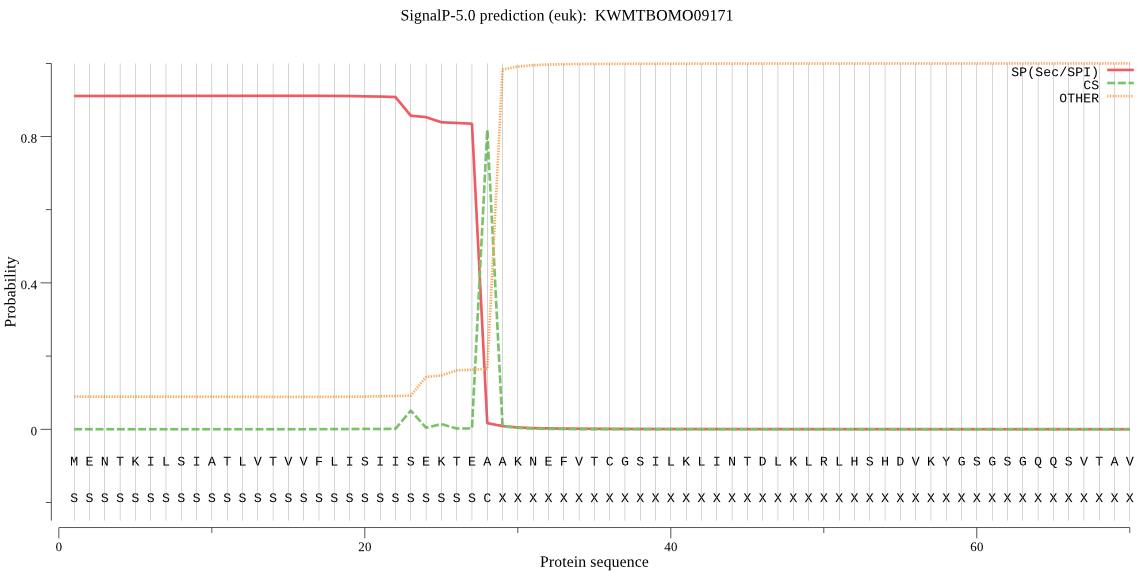
SignalP
Position: 1 - 28,
Likelihood: 0.911355
Length:
224
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.95706
Exp number, first 60 AAs:
11.95683
Total prob of N-in:
0.43286
POSSIBLE N-term signal
sequence
outside
1 - 224
Population Genetic Test Statistics
Pi
187.951377
Theta
167.866448
Tajima's D
0.500233
CLR
0.064188
CSRT
0.511174441277936
Interpretation
Uncertain
