Gene
KWMTBOMO09162
Pre Gene Modal
BGIBMGA003372
Annotation
PREDICTED:_tigger_transposable_element-derived_protein_6-like_isoform_X3_[Pogonomyrmex_barbatus]
Location in the cell
Nuclear Reliability : 3.045
Sequence
CDS
ATGAGGACATATAAAAGAAAAACGGAGCGTGGCAAAATATCAAAAGATGTTTACGAACAAGCCGCGGTTATATTGGAGGAAGATGAGTCAAAAAAAATTCGTGGCATCGCAAAAGATTTCGGATTATGTCATATGTCGCTAACGAGATATATGAGGAAAAGTAAAGATGCAAAACAGAAGGGTATACCTATAGAATCACTCACAATAGGATACAAGAAGAATCGTCAAGTATTCAGTGGTGAACAGGAAGCAATTTTAGTGAGTTATTTGATTAAATGTAGCCAAATTTATTACGGCTTAACTCCAAAAGACGTCAGACAACTTGCATTTGATTGCGCACTAAAGCACAATATTGCGATACCAAAATCGTGGCTTGACAATAAAGAGGCAGGTGTAGATTGGCTGACAGCGTTTTTAAAACGCAACCCGTCTTTATCTATACGGTCGCCAGAAGCTACAAGCTTGAGCAGAGCCACATCATTCAATAAAACGAATGTTGACAATTTCTTTTCTAAGTTGACTGACGTGTTAGACAGATATAGATTTTCTGCATCACGTATATGGAATGTGGACGAGACAGGTGTCACCACGGTGCTGAAACCCAGAAAAATTTTAGCTGCAAAGGGTTCTAAACAAGTTGGTTCAATTACATCAGCCGAGAGAGGGACTCTTGTGACTCTCTGCGTCGCAGTAAATGCAGTGGGAAATTCGGTGCCTTGTATGTTCATCTTTCCACGAATTAAATATAGAGAATATTTTGTTCGCGATGGACCACCTGGTTCAATTGGTGCAGGCAACTCAAGTGGCTGGATGACAGGGCGAGAATTTAAAATTTTTATGAAACATTTTATAGATAATGTGAAACCATCTCCATCCGATCCTGTGCTCTTATTATTAGACAATCATACTTCGCATTTGGATATTGAAGTCATTGAAACAGCCAAAGACAATAATGTAGTGTTACTCTCCTTTCCACCACACTGCTCACATAAGCTCCAGCCCTTGGATGTGGGGGTTTATGGTCCATTAAAAAATTACTGTGCCAGTCAGCAAGATTCTTGGCTTAGAAACAATCCAGGGAAAACAATGTCTATTCATGACATTCCATCTATAGTGAATAAAGCTTTACCCTTAGCATTGAATCCAGCTAACATTATCAATGGATTTAAAAATACTGGCATCGCACCATTTAATAAAGATATATTCCAGGAAGATGACTTCTTATCGGCTTTTGTAACTGATAGACCAATAGACGACGAAGTGCCTAATGCAAACATTGAAAGTTGCCCTGTTACAACGCAGAATAAAGAGACAGAAACTCCAGTTTCACCTTCTTGCCATCGCCCCATAATAAGCCCTATAACAGAAGCCAATAGGTCAATTTATACCAGCCCAGTTGCGGGATGTTCTAAAGACATTGAAAATACTTTTTCTCCTGCAAATATTCGACCTTATCCAAAAGCAGCTGCCAGAAAAAGCAAATTAACTCGGAAAACTCGCAAATCCGCTATTTTAACTGATACACCGGAAAAAAATGCTTTGGCTTTGGAACAAGAGAAAAGGAACGATATTAAATCTAAAAAGGGAAAGTTAACTAAAAGATTAAAGTCAAAATCAAGTTCCGAGAAGACAGATAAACAAAATGTGAAGAAAATAAGAAAAACAATTAAGCAAATAAAACAGTTAGTAGTTAAGAAGAAGATATTACATGAAACTGATGAAGAAATCGTAGAAGATGAATATTTTTGTCTGATATGTTGTGACCCATTTGATCCCAAGAAAGCTGGTGAGGAATGGATAGAGTGTATTGTTTGCAAAAATTGGTCACATGTGAAATGTATCCGAGGAAATATTATACAATACGTTTGTTTAAATTGCGAAAGTGACCATTCAGATTACAGTGACTAA
Protein
MRTYKRKTERGKISKDVYEQAAVILEEDESKKIRGIAKDFGLCHMSLTRYMRKSKDAKQKGIPIESLTIGYKKNRQVFSGEQEAILVSYLIKCSQIYYGLTPKDVRQLAFDCALKHNIAIPKSWLDNKEAGVDWLTAFLKRNPSLSIRSPEATSLSRATSFNKTNVDNFFSKLTDVLDRYRFSASRIWNVDETGVTTVLKPRKILAAKGSKQVGSITSAERGTLVTLCVAVNAVGNSVPCMFIFPRIKYREYFVRDGPPGSIGAGNSSGWMTGREFKIFMKHFIDNVKPSPSDPVLLLLDNHTSHLDIEVIETAKDNNVVLLSFPPHCSHKLQPLDVGVYGPLKNYCASQQDSWLRNNPGKTMSIHDIPSIVNKALPLALNPANIINGFKNTGIAPFNKDIFQEDDFLSAFVTDRPIDDEVPNANIESCPVTTQNKETETPVSPSCHRPIISPITEANRSIYTSPVAGCSKDIENTFSPANIRPYPKAAARKSKLTRKTRKSAILTDTPEKNALALEQEKRNDIKSKKGKLTKRLKSKSSSEKTDKQNVKKIRKTIKQIKQLVVKKKILHETDEEIVEDEYFCLICCDPFDPKKAGEEWIECIVCKNWSHVKCIRGNIIQYVCLNCESDHSDYSD
Summary
Uniprot
A0A1W4WAP5
A0A2A4JQE1
A0A151IRD9
A0A2A4J1E0
A0A1W4XA29
A0A2S2NP39
+ More
A0A2A4K4E0 A0A2H1VA32 A0A151IB56 A0A3N0Y1T9 A0A0J7KEH1 A0A2W1B5C9 X1WJ54 A0A0P6I744 A0A0P5JU33 A0A1Y1N139 A0A1Y1MZ46 X1XKW0 X1XDD3 A0A2L2YKN8 J9K539 A0A2H1W686 A0A0P5M4C5 A0A3P8NPX0 A0A147BB25 A0A164L4P2 J9K2D5 A0A0P5JXG0 A0A0P6HDU5 X1WM14 A0A146KPT3 A0A3P8XDA2 L7MAV6 A0A1Y1L3N3 A0A1Y1MAV5 A0A151X7P4 A0A0P5Q3E2 E9H8A3 A0A2H1VRL1 J9KKA5 A0A1B6L3T1 A0A0P5BKE5
A0A2A4K4E0 A0A2H1VA32 A0A151IB56 A0A3N0Y1T9 A0A0J7KEH1 A0A2W1B5C9 X1WJ54 A0A0P6I744 A0A0P5JU33 A0A1Y1N139 A0A1Y1MZ46 X1XKW0 X1XDD3 A0A2L2YKN8 J9K539 A0A2H1W686 A0A0P5M4C5 A0A3P8NPX0 A0A147BB25 A0A164L4P2 J9K2D5 A0A0P5JXG0 A0A0P6HDU5 X1WM14 A0A146KPT3 A0A3P8XDA2 L7MAV6 A0A1Y1L3N3 A0A1Y1MAV5 A0A151X7P4 A0A0P5Q3E2 E9H8A3 A0A2H1VRL1 J9KKA5 A0A1B6L3T1 A0A0P5BKE5
EMBL
NWSH01000827
PCG73986.1
KQ981133
KYN09256.1
NWSH01003711
PCG65895.1
+ More
GGMR01006341 MBY18960.1 NWSH01000193 PCG78522.1 ODYU01001473 SOQ37710.1 KQ978123 KYM96896.1 RJVU01053773 ROL23530.1 LBMM01008804 KMQ88629.1 KZ150321 PZC71419.1 ABLF02029656 GDIQ01010354 JAN84383.1 GDIQ01193792 JAK57933.1 GEZM01020578 JAV89247.1 GEZM01020579 JAV89246.1 ABLF02018523 ABLF02018527 ABLF02023249 ABLF02023256 IAAA01031058 LAA08688.1 ABLF02011603 ODYU01006608 SOQ48601.1 GDIQ01164503 JAK87222.1 GEGO01007436 JAR87968.1 LRGB01003206 KZS03791.1 ABLF02010860 ABLF02010863 GDIQ01193793 JAK57932.1 GDIQ01020929 JAN73808.1 ABLF02007358 ABLF02007359 ABLF02007364 ABLF02037183 GDHC01020924 JAP97704.1 GACK01003884 JAA61150.1 GEZM01066092 GEZM01066091 JAV68194.1 GEZM01039017 JAV81445.1 KQ982443 KYQ56383.1 GDIQ01126265 JAL25461.1 GL732603 EFX72035.1 ODYU01004023 SOQ43475.1 ABLF02006839 GEBQ01021595 JAT18382.1 GDIP01183765 JAJ39637.1
GGMR01006341 MBY18960.1 NWSH01000193 PCG78522.1 ODYU01001473 SOQ37710.1 KQ978123 KYM96896.1 RJVU01053773 ROL23530.1 LBMM01008804 KMQ88629.1 KZ150321 PZC71419.1 ABLF02029656 GDIQ01010354 JAN84383.1 GDIQ01193792 JAK57933.1 GEZM01020578 JAV89247.1 GEZM01020579 JAV89246.1 ABLF02018523 ABLF02018527 ABLF02023249 ABLF02023256 IAAA01031058 LAA08688.1 ABLF02011603 ODYU01006608 SOQ48601.1 GDIQ01164503 JAK87222.1 GEGO01007436 JAR87968.1 LRGB01003206 KZS03791.1 ABLF02010860 ABLF02010863 GDIQ01193793 JAK57932.1 GDIQ01020929 JAN73808.1 ABLF02007358 ABLF02007359 ABLF02007364 ABLF02037183 GDHC01020924 JAP97704.1 GACK01003884 JAA61150.1 GEZM01066092 GEZM01066091 JAV68194.1 GEZM01039017 JAV81445.1 KQ982443 KYQ56383.1 GDIQ01126265 JAL25461.1 GL732603 EFX72035.1 ODYU01004023 SOQ43475.1 ABLF02006839 GEBQ01021595 JAT18382.1 GDIP01183765 JAJ39637.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A1W4WAP5
A0A2A4JQE1
A0A151IRD9
A0A2A4J1E0
A0A1W4XA29
A0A2S2NP39
+ More
A0A2A4K4E0 A0A2H1VA32 A0A151IB56 A0A3N0Y1T9 A0A0J7KEH1 A0A2W1B5C9 X1WJ54 A0A0P6I744 A0A0P5JU33 A0A1Y1N139 A0A1Y1MZ46 X1XKW0 X1XDD3 A0A2L2YKN8 J9K539 A0A2H1W686 A0A0P5M4C5 A0A3P8NPX0 A0A147BB25 A0A164L4P2 J9K2D5 A0A0P5JXG0 A0A0P6HDU5 X1WM14 A0A146KPT3 A0A3P8XDA2 L7MAV6 A0A1Y1L3N3 A0A1Y1MAV5 A0A151X7P4 A0A0P5Q3E2 E9H8A3 A0A2H1VRL1 J9KKA5 A0A1B6L3T1 A0A0P5BKE5
A0A2A4K4E0 A0A2H1VA32 A0A151IB56 A0A3N0Y1T9 A0A0J7KEH1 A0A2W1B5C9 X1WJ54 A0A0P6I744 A0A0P5JU33 A0A1Y1N139 A0A1Y1MZ46 X1XKW0 X1XDD3 A0A2L2YKN8 J9K539 A0A2H1W686 A0A0P5M4C5 A0A3P8NPX0 A0A147BB25 A0A164L4P2 J9K2D5 A0A0P5JXG0 A0A0P6HDU5 X1WM14 A0A146KPT3 A0A3P8XDA2 L7MAV6 A0A1Y1L3N3 A0A1Y1MAV5 A0A151X7P4 A0A0P5Q3E2 E9H8A3 A0A2H1VRL1 J9KKA5 A0A1B6L3T1 A0A0P5BKE5
Ontologies
GO
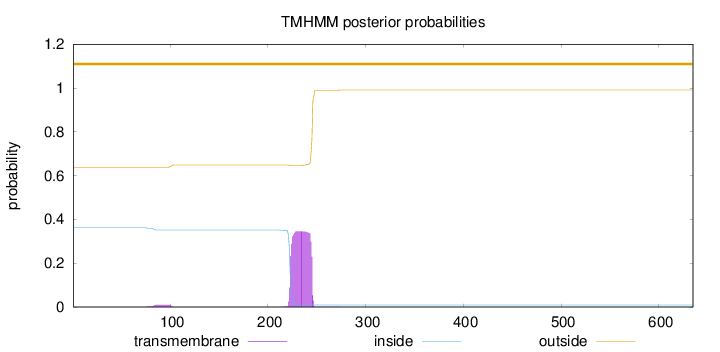
Topology
Subcellular location
Nucleus
Length:
635
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.95834999999999
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.36149
outside
1 - 635
Population Genetic Test Statistics
Pi
151.115543
Theta
170.627656
Tajima's D
1.080577
CLR
0
CSRT
0.684815759212039
Interpretation
Uncertain
