Gene
KWMTBOMO09136 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007553
Annotation
C-type_lectin_16_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.965
Sequence
CDS
ATGAAGACATCAGTACTCGTATTAGCTGCGCTAGCGGTGGTGGCCCAGGCACAGTTCCCCAATGGACGCATCCTGGAGCCTCCGGTGCCTTCTTTGTGTGTACAGAGAAACATCCATGAGAGATACGCTGACAATAAAGGATACTTCTTCTCGTGGCGGGACCCAGCGCTCCGCGGCGTCGAAGAAGACTGGTTGAGCGCAAGAAACTACTGCCGACAACGCTGCATGGACCTAGTATCTCTCGAGACCAGCGATGAAAATGAATGGGTCAAAGCGCGTATTGTCCAGGACAAGATAAAGTACATTTGGACATCGGGCCGTCTTTGTGACTTCAAAGGATGCAACCGACCTGACTTACTACCTAACGAAATAAACGGTTGGTTCTGGACTGCTGAGCTGCAGAAGTTGGCTCCCACCACCAACCGTCAACAGAACGACTGGTCCGAAGGTGGCGGCATTGGAAAACCGCAACCTGATAACCGGGAGCTGATACAAGGAGGCGCTTCTGAGCACTGCGTCGCAATCTTAAACAACTTCTACAACGACGGTGTCCACTGGCACGACGTCGCCTGCCACCACCGCAAGCCATTCGTTTGCGAAGAAAACGACGCCCTCCTCAAATACGTCAGATATACCAATCCTAACTTAAGAGTTTAA
Protein
MKTSVLVLAALAVVAQAQFPNGRILEPPVPSLCVQRNIHERYADNKGYFFSWRDPALRGVEEDWLSARNYCRQRCMDLVSLETSDENEWVKARIVQDKIKYIWTSGRLCDFKGCNRPDLLPNEINGWFWTAELQKLAPTTNRQQNDWSEGGGIGKPQPDNRELIQGGASEHCVAILNNFYNDGVHWHDVACHHRKPFVCEENDALLKYVRYTNPNLRV
Summary
Uniprot
Q06FJ6
H9JDF6
A0A3S2NBT5
C5MKD8
A0A2A4JG01
A0A3G1HH33
+ More
A0A194PJI4 A0A2H1VTL1 A0A194R7W3 I4DIW7 I4DM01 A0A212ENZ7 A0A0L7LKW9 A0A219YXG3 A0A336LMG5 A0A1W4WHX0 D6WDS2 A0A182X8X5 A0A453Z1Z7 A0A182I407 A0A182WC61 A0A182PID1 A0A182YP87 A0A182MT07 A0A182LL24 A0A0T6BGK5 A0A2Z5U8B4 A0A182FEZ6 A0A182QLT4 A0NET0 A0A182IMV9 A0A1J1HZE0 B0WCD7 A0A182NJ63 A0A1Q3FPK1 K7J8I6 A0A232FBA3 Q27U52 T1PIW2 Q16HZ2 A0A182GV37 A0A0K8U027 A0A034WGB5 A0A0L0CLR7 A0A0A1XNU4 A0A1I8NX82 E2BE27 A0A0M3QXR0 W8C1U0 A0A2P8YH27 B4K9E8 B4N819 B3LZK9 B4LXK9 A0A026WID6 B3P1D5 B4JTJ5 A0A3B0JTP9 Q295Q6 B4GFU6 A0A067QXP7 B4QSY0 B4HHE3 Q9VGA3 A0A195F1W0 B4PNQ7 A0A195E6H3 A0A195B1S9 A0A158NLD6 A0A195CY78 E2ATA6 F4X6L1 A0A171AVG9 A0A1W4UBI5 A0A2J7PRS1 A0A1V1G0Z1 A0A151XCB4 A0A1V1G4Y2 A0A087ZXS4 A0A2A3E3G0 E9IZ65 T1H8Y7 A0A1B6HNC4 A0A0L7R4U9 A0A1B6CVR4 A0A146L1I4 A0A0A9W0J6 N6U9S1 A0A310SLC1 A0A154PGK1 A0A0N0BH81 U4UBI5 A0A0K8S2Z9 N6UNW5 U4U2V5 A0A2P8Y7L9 A0A067RBR3 A0A1V1FIP4 A0A2H1X2K3
A0A194PJI4 A0A2H1VTL1 A0A194R7W3 I4DIW7 I4DM01 A0A212ENZ7 A0A0L7LKW9 A0A219YXG3 A0A336LMG5 A0A1W4WHX0 D6WDS2 A0A182X8X5 A0A453Z1Z7 A0A182I407 A0A182WC61 A0A182PID1 A0A182YP87 A0A182MT07 A0A182LL24 A0A0T6BGK5 A0A2Z5U8B4 A0A182FEZ6 A0A182QLT4 A0NET0 A0A182IMV9 A0A1J1HZE0 B0WCD7 A0A182NJ63 A0A1Q3FPK1 K7J8I6 A0A232FBA3 Q27U52 T1PIW2 Q16HZ2 A0A182GV37 A0A0K8U027 A0A034WGB5 A0A0L0CLR7 A0A0A1XNU4 A0A1I8NX82 E2BE27 A0A0M3QXR0 W8C1U0 A0A2P8YH27 B4K9E8 B4N819 B3LZK9 B4LXK9 A0A026WID6 B3P1D5 B4JTJ5 A0A3B0JTP9 Q295Q6 B4GFU6 A0A067QXP7 B4QSY0 B4HHE3 Q9VGA3 A0A195F1W0 B4PNQ7 A0A195E6H3 A0A195B1S9 A0A158NLD6 A0A195CY78 E2ATA6 F4X6L1 A0A171AVG9 A0A1W4UBI5 A0A2J7PRS1 A0A1V1G0Z1 A0A151XCB4 A0A1V1G4Y2 A0A087ZXS4 A0A2A3E3G0 E9IZ65 T1H8Y7 A0A1B6HNC4 A0A0L7R4U9 A0A1B6CVR4 A0A146L1I4 A0A0A9W0J6 N6U9S1 A0A310SLC1 A0A154PGK1 A0A0N0BH81 U4UBI5 A0A0K8S2Z9 N6UNW5 U4U2V5 A0A2P8Y7L9 A0A067RBR3 A0A1V1FIP4 A0A2H1X2K3
Pubmed
19121390
19442669
26354079
22651552
22118469
26227816
+ More
18362917 19820115 25244985 20966253 12364791 14747013 17210077 20075255 28648823 16907828 25315136 17510324 26483478 25348373 26108605 25830018 20798317 24495485 29403074 17994087 24508170 30249741 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21347285 21719571 28410430 21282665 26823975 25401762 23537049
18362917 19820115 25244985 20966253 12364791 14747013 17210077 20075255 28648823 16907828 25315136 17510324 26483478 25348373 26108605 25830018 20798317 24495485 29403074 17994087 24508170 30249741 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21347285 21719571 28410430 21282665 26823975 25401762 23537049
EMBL
DQ898219
ABI79325.1
BABH01030069
RSAL01000115
RVE46913.1
GQ120500
+ More
ACR78452.1 NWSH01001662 PCG70494.1 KY111306 AQX37242.1 KQ459602 KPI93487.1 ODYU01004370 SOQ44179.1 KQ460615 KPJ13737.1 AK401235 BAM17857.1 AK402319 BAM18941.1 AGBW02013545 OWR43225.1 JTDY01000728 KOB76087.1 KX550114 AQY54431.1 UFQS01000016 UFQT01000016 SSW97457.1 SSX17843.1 KQ971322 EFA01339.2 APCN01002263 APCN01002264 AXCM01002383 AXCM01002384 LJIG01000514 KRT86426.1 AP017517 BBB06860.1 AXCN02000455 AAAB01008960 EAU76601.1 CVRI01000035 CRK92692.1 DS231886 EDS43382.1 GFDL01005515 JAV29530.1 NNAY01000478 OXU28106.1 DQ377067 ABD60991.1 KA648614 AFP63243.1 CH478126 CH478056 EAT33884.1 EAT34165.1 EJY58059.1 JXUM01090266 JXUM01090267 JXUM01090268 KQ563828 KXJ73210.1 GDHF01032441 JAI19873.1 GAKP01005243 GAKP01005242 JAC53709.1 JRES01000223 KNC33206.1 GBXI01001675 JAD12617.1 GL447755 EFN86014.1 CP012526 ALC46363.1 GAMC01003407 JAC03149.1 PYGN01000600 PSN43551.1 CH933806 EDW15580.1 CH964232 EDW81270.1 CH902617 EDV41951.1 CH940650 EDW66793.1 KK107185 QOIP01000012 EZA55812.1 RLU15747.1 CH954181 EDV49394.1 CH916373 EDV95085.1 OUUW01000008 SPP84433.1 CM000070 EAL28652.1 CH479182 EDW34481.1 KK853131 KDR10900.1 CM000364 EDX13240.1 CH480815 EDW42482.1 AE014297 BT030988 AAF54782.2 ABV82370.1 KQ981864 KYN34366.1 CM000160 EDW97072.1 KQ979568 KYN20773.1 KQ976662 KYM78428.1 ADTU01019320 KQ977115 KYN05608.1 GL442545 EFN63351.1 GL888818 EGI57793.1 GEMB01000604 JAS02528.1 NEVH01021953 PNF19019.1 FX985244 BAX07257.1 KQ982314 KYQ58021.1 FX985243 BAX07256.1 KZ288430 PBC25832.1 GL767119 EFZ14123.1 ACPB03000427 GECU01031519 JAS76187.1 KQ414657 KOC65801.1 GEDC01019781 JAS17517.1 GDHC01016215 JAQ02414.1 GBHO01042245 JAG01359.1 APGK01034109 KB740904 ENN78440.1 KQ760118 OAD61924.1 KQ434893 KZC10594.1 KQ435757 KOX75842.1 KB632006 ERL87951.1 GBRD01018165 JAG47662.1 APGK01003087 APGK01018482 APGK01018483 KB740076 KB735111 ENN81657.1 ENN83445.1 KB631643 ERL84921.1 PYGN01000830 PSN40257.1 KK852561 KDR21331.1 FX985238 BAX07251.1 ODYU01012907 SOQ59446.1
ACR78452.1 NWSH01001662 PCG70494.1 KY111306 AQX37242.1 KQ459602 KPI93487.1 ODYU01004370 SOQ44179.1 KQ460615 KPJ13737.1 AK401235 BAM17857.1 AK402319 BAM18941.1 AGBW02013545 OWR43225.1 JTDY01000728 KOB76087.1 KX550114 AQY54431.1 UFQS01000016 UFQT01000016 SSW97457.1 SSX17843.1 KQ971322 EFA01339.2 APCN01002263 APCN01002264 AXCM01002383 AXCM01002384 LJIG01000514 KRT86426.1 AP017517 BBB06860.1 AXCN02000455 AAAB01008960 EAU76601.1 CVRI01000035 CRK92692.1 DS231886 EDS43382.1 GFDL01005515 JAV29530.1 NNAY01000478 OXU28106.1 DQ377067 ABD60991.1 KA648614 AFP63243.1 CH478126 CH478056 EAT33884.1 EAT34165.1 EJY58059.1 JXUM01090266 JXUM01090267 JXUM01090268 KQ563828 KXJ73210.1 GDHF01032441 JAI19873.1 GAKP01005243 GAKP01005242 JAC53709.1 JRES01000223 KNC33206.1 GBXI01001675 JAD12617.1 GL447755 EFN86014.1 CP012526 ALC46363.1 GAMC01003407 JAC03149.1 PYGN01000600 PSN43551.1 CH933806 EDW15580.1 CH964232 EDW81270.1 CH902617 EDV41951.1 CH940650 EDW66793.1 KK107185 QOIP01000012 EZA55812.1 RLU15747.1 CH954181 EDV49394.1 CH916373 EDV95085.1 OUUW01000008 SPP84433.1 CM000070 EAL28652.1 CH479182 EDW34481.1 KK853131 KDR10900.1 CM000364 EDX13240.1 CH480815 EDW42482.1 AE014297 BT030988 AAF54782.2 ABV82370.1 KQ981864 KYN34366.1 CM000160 EDW97072.1 KQ979568 KYN20773.1 KQ976662 KYM78428.1 ADTU01019320 KQ977115 KYN05608.1 GL442545 EFN63351.1 GL888818 EGI57793.1 GEMB01000604 JAS02528.1 NEVH01021953 PNF19019.1 FX985244 BAX07257.1 KQ982314 KYQ58021.1 FX985243 BAX07256.1 KZ288430 PBC25832.1 GL767119 EFZ14123.1 ACPB03000427 GECU01031519 JAS76187.1 KQ414657 KOC65801.1 GEDC01019781 JAS17517.1 GDHC01016215 JAQ02414.1 GBHO01042245 JAG01359.1 APGK01034109 KB740904 ENN78440.1 KQ760118 OAD61924.1 KQ434893 KZC10594.1 KQ435757 KOX75842.1 KB632006 ERL87951.1 GBRD01018165 JAG47662.1 APGK01003087 APGK01018482 APGK01018483 KB740076 KB735111 ENN81657.1 ENN83445.1 KB631643 ERL84921.1 PYGN01000830 PSN40257.1 KK852561 KDR21331.1 FX985238 BAX07251.1 ODYU01012907 SOQ59446.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000192223 UP000007266 UP000076407 UP000075903 UP000075840 UP000075920 UP000075885 UP000076408 UP000075883 UP000075882 UP000069272 UP000075886 UP000007062 UP000075880 UP000183832 UP000002320 UP000075884 UP000002358 UP000215335 UP000095301 UP000008820 UP000069940 UP000249989 UP000037069 UP000095300 UP000008237 UP000092553 UP000245037 UP000009192 UP000007798 UP000007801 UP000008792 UP000053097 UP000279307 UP000008711 UP000001070 UP000268350 UP000001819 UP000008744 UP000027135 UP000000304 UP000001292 UP000000803 UP000078541 UP000002282 UP000078492 UP000078540 UP000005205 UP000078542 UP000000311 UP000007755 UP000192221 UP000235965 UP000075809 UP000005203 UP000242457 UP000015103 UP000053825 UP000019118 UP000076502 UP000053105 UP000030742
UP000037510 UP000192223 UP000007266 UP000076407 UP000075903 UP000075840 UP000075920 UP000075885 UP000076408 UP000075883 UP000075882 UP000069272 UP000075886 UP000007062 UP000075880 UP000183832 UP000002320 UP000075884 UP000002358 UP000215335 UP000095301 UP000008820 UP000069940 UP000249989 UP000037069 UP000095300 UP000008237 UP000092553 UP000245037 UP000009192 UP000007798 UP000007801 UP000008792 UP000053097 UP000279307 UP000008711 UP000001070 UP000268350 UP000001819 UP000008744 UP000027135 UP000000304 UP000001292 UP000000803 UP000078541 UP000002282 UP000078492 UP000078540 UP000005205 UP000078542 UP000000311 UP000007755 UP000192221 UP000235965 UP000075809 UP000005203 UP000242457 UP000015103 UP000053825 UP000019118 UP000076502 UP000053105 UP000030742
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
Q06FJ6
H9JDF6
A0A3S2NBT5
C5MKD8
A0A2A4JG01
A0A3G1HH33
+ More
A0A194PJI4 A0A2H1VTL1 A0A194R7W3 I4DIW7 I4DM01 A0A212ENZ7 A0A0L7LKW9 A0A219YXG3 A0A336LMG5 A0A1W4WHX0 D6WDS2 A0A182X8X5 A0A453Z1Z7 A0A182I407 A0A182WC61 A0A182PID1 A0A182YP87 A0A182MT07 A0A182LL24 A0A0T6BGK5 A0A2Z5U8B4 A0A182FEZ6 A0A182QLT4 A0NET0 A0A182IMV9 A0A1J1HZE0 B0WCD7 A0A182NJ63 A0A1Q3FPK1 K7J8I6 A0A232FBA3 Q27U52 T1PIW2 Q16HZ2 A0A182GV37 A0A0K8U027 A0A034WGB5 A0A0L0CLR7 A0A0A1XNU4 A0A1I8NX82 E2BE27 A0A0M3QXR0 W8C1U0 A0A2P8YH27 B4K9E8 B4N819 B3LZK9 B4LXK9 A0A026WID6 B3P1D5 B4JTJ5 A0A3B0JTP9 Q295Q6 B4GFU6 A0A067QXP7 B4QSY0 B4HHE3 Q9VGA3 A0A195F1W0 B4PNQ7 A0A195E6H3 A0A195B1S9 A0A158NLD6 A0A195CY78 E2ATA6 F4X6L1 A0A171AVG9 A0A1W4UBI5 A0A2J7PRS1 A0A1V1G0Z1 A0A151XCB4 A0A1V1G4Y2 A0A087ZXS4 A0A2A3E3G0 E9IZ65 T1H8Y7 A0A1B6HNC4 A0A0L7R4U9 A0A1B6CVR4 A0A146L1I4 A0A0A9W0J6 N6U9S1 A0A310SLC1 A0A154PGK1 A0A0N0BH81 U4UBI5 A0A0K8S2Z9 N6UNW5 U4U2V5 A0A2P8Y7L9 A0A067RBR3 A0A1V1FIP4 A0A2H1X2K3
A0A194PJI4 A0A2H1VTL1 A0A194R7W3 I4DIW7 I4DM01 A0A212ENZ7 A0A0L7LKW9 A0A219YXG3 A0A336LMG5 A0A1W4WHX0 D6WDS2 A0A182X8X5 A0A453Z1Z7 A0A182I407 A0A182WC61 A0A182PID1 A0A182YP87 A0A182MT07 A0A182LL24 A0A0T6BGK5 A0A2Z5U8B4 A0A182FEZ6 A0A182QLT4 A0NET0 A0A182IMV9 A0A1J1HZE0 B0WCD7 A0A182NJ63 A0A1Q3FPK1 K7J8I6 A0A232FBA3 Q27U52 T1PIW2 Q16HZ2 A0A182GV37 A0A0K8U027 A0A034WGB5 A0A0L0CLR7 A0A0A1XNU4 A0A1I8NX82 E2BE27 A0A0M3QXR0 W8C1U0 A0A2P8YH27 B4K9E8 B4N819 B3LZK9 B4LXK9 A0A026WID6 B3P1D5 B4JTJ5 A0A3B0JTP9 Q295Q6 B4GFU6 A0A067QXP7 B4QSY0 B4HHE3 Q9VGA3 A0A195F1W0 B4PNQ7 A0A195E6H3 A0A195B1S9 A0A158NLD6 A0A195CY78 E2ATA6 F4X6L1 A0A171AVG9 A0A1W4UBI5 A0A2J7PRS1 A0A1V1G0Z1 A0A151XCB4 A0A1V1G4Y2 A0A087ZXS4 A0A2A3E3G0 E9IZ65 T1H8Y7 A0A1B6HNC4 A0A0L7R4U9 A0A1B6CVR4 A0A146L1I4 A0A0A9W0J6 N6U9S1 A0A310SLC1 A0A154PGK1 A0A0N0BH81 U4UBI5 A0A0K8S2Z9 N6UNW5 U4U2V5 A0A2P8Y7L9 A0A067RBR3 A0A1V1FIP4 A0A2H1X2K3
PDB
5VC1
E-value=0.0119463,
Score=87
Ontologies
Topology
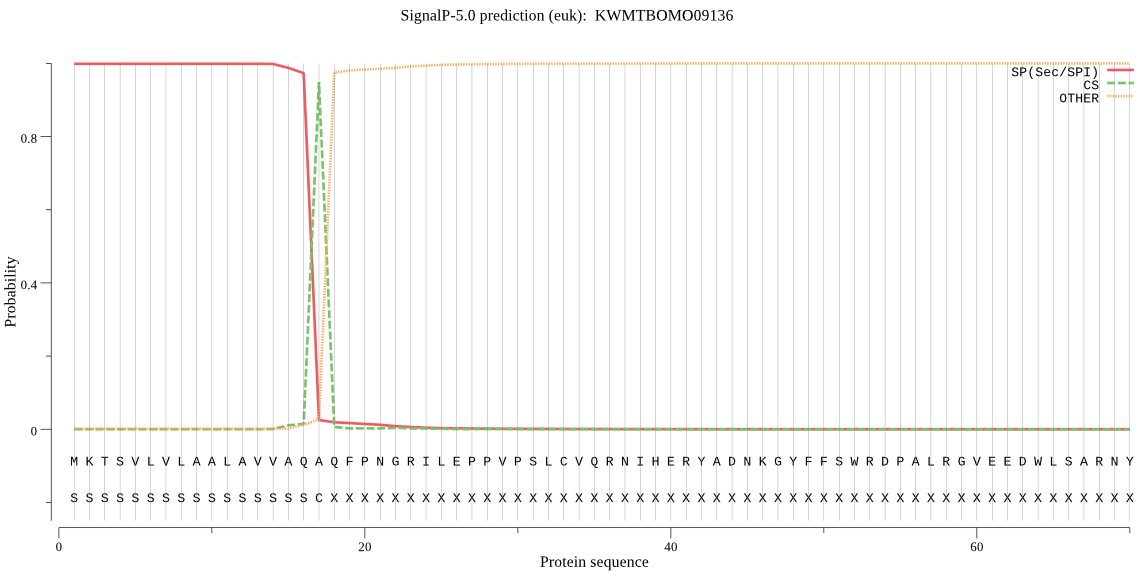
SignalP
Position: 1 - 17,
Likelihood: 0.998596
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44215
Exp number, first 60 AAs:
0.44215
Total prob of N-in:
0.07732
outside
1 - 218

Population Genetic Test Statistics
Pi
262.123458
Theta
185.035012
Tajima's D
1.18638
CLR
1.411187
CSRT
0.710464476776161
Interpretation
Uncertain
