Gene
KWMTBOMO09132
Pre Gene Modal
BGIBMGA007542
Annotation
PREDICTED:_uncharacterized_protein_LOC106136994_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.134
Sequence
CDS
ATGTCGTTTTTTAAGAGCGTCGCTTCTGGATTCACAACACGCAGTCGTGACTCAATTCTTAAAGAATCAATAATAAACAACTTATCGGAAAGTGTTAAAGACATACAGTATAACAATCAATTTGAAGAAAATTTTCACAGTGTTCTTGGTTCGACCGATTCTACAAGTACCCTGTGCATGGCCCTTGAAGCAGTCTTTCTTCACGGTCTCAAGGACACATTATTAAGAAAAGCCAAAAATGCTCTCACTGGCGATTCTGATCAGCAACCCGAGCCAAACTTTTGGCCGCTCGTTCTAGTACTGTCTCACAGACAAAATATAGATCAAATTTCATCTTTGCCTCAGATCAACACAGAAATTGGTCAGTGCCGAGCTTGGCTGCGTATTGCTTTGAATGAATGTTTGCTGTCCTCATATATGTCAACTCTACACAAAAATATTTCAGCTATAAAACCATTCTACAATCGTAATGCATTAGTATGTGATCCTGAGATATTGGATGTGGCACAAAGGCTAGTAGAAGGTCTAGAGACATGTGTACAGTTCAATTTACCGATTAATTCAAGTCTATTAAATTACTGGCCTGAACAAGTATTAATAATAGCAGGTATTTGGGTGCCCAGGGTGAGGGTTGCTCCCATCAGTGCAGCATTAGATGTGGCAAGTATTATCACTGAAGAAGAAGATAAACCTAAATTGGCATCTACCCCTATACACACACCTAGCATCACTGGGTTGGGTCCAATACTTGCTCTCAATGAGGATGAAGCTTTGGATTATATTCTATCTAAAAATTCAAGTGATATTAGAAAAGTAACAAAGAGTAATTTGGATGAAAATGACAAAAATGAAACTATTGAAAGTAAAAATGCAGATGTTATATGTGATGAAATGGCTACTAATGATGGATCATTACCACTTCCAATACAATATCCATCAACAAGTCAAGATGCAGCAGATGAACTGCTTGCCTCCCCTACCTCAGTGAAATCTGAAGATTTTGAAGCTCATTGTGAAGTTGTTATTACTGGCAACTCATTAATTGGTAAAGGTTGGTCAGCAGATAATAATGATGATTTAAATGCATCAATTCATTCAAATTCCTCACATGGCAGTAGTATGATTAAAACTCCAGAAATAAAAAGTCTGTCTTCATTAGTGGACCAAACAAATCCTTTTGCTGAAACATTCTCTAACATGCCAAATCTGAGAGACATCATCAAGGATATACATAGAGAACTGAAGTTTACTCCAGATGATGTTGCTAGAAGTAACATAAAAATTGATCCTGTTACACCAGAAGACCCTGCTCTTCAGGGTTTGGACTTTGAAGTTGTTCCAAGTTCAATGACAGAATCATTTACTGTTCCTGAACTACAGAGCTTGATACAACAGTTGGGGGTACTATCAAGAGAACAGGGTCTTGATCATCAAAACTACCAGTGCAAAGGGTGTCAGGATTTGTTGGGAAGTACAATATCAAGAGCTAAAGTTTGTGCTTACACTGGAGAATACTATTGCTCTAATTGCATGGATCCAAATGTTTTTATTATTCCTGCCAGAGTTATACATAATTGGGATTTCAAAAGATACCCAGTTTCTAAAAAAGCAGCCCTGTTCTTATTAGAATTTCAACATCATCCATGGATAGATATGAAGAAATTAAATCCTAAAATATACTGCGGAGTTTCAGATATGGCCCATTTACAAGATCTTCGAATACAACTGAACTTTTTAAGGGCTTACATTTTTACATGTCGGGAGCCTGTTATTGAAGATCTGCAGAAACGTATATGGCCAAGAGAACACTTGTATGAGCATGTCCATTTGTATACTATTTCAGATTTGGCACAGATTCCAAATGGTTCCTTGGTTTTGCAGTTAGAAAAAGTGGTTTCATTTGCTAGGTCTCATGTATTGGACTGTTGGCTTTGTAGTCAGAAAGGTTTCATTTGCGAGGTTTGCAAAGATCCAAAAATTTTGTATCCATTCGAAACTGCCGGTACATACAGATGTGACGAATGTTCCAGTGTATTTCATGACAGATGTTTGAATGTAAATATACCATGTCCAAAATGTAAGAGGAAGCAGAATCGCACAAATGACACATCATTAGTTGATGCTGTAAATGCTTGTTTCAACAAATTATAA
Protein
MSFFKSVASGFTTRSRDSILKESIINNLSESVKDIQYNNQFEENFHSVLGSTDSTSTLCMALEAVFLHGLKDTLLRKAKNALTGDSDQQPEPNFWPLVLVLSHRQNIDQISSLPQINTEIGQCRAWLRIALNECLLSSYMSTLHKNISAIKPFYNRNALVCDPEILDVAQRLVEGLETCVQFNLPINSSLLNYWPEQVLIIAGIWVPRVRVAPISAALDVASIITEEEDKPKLASTPIHTPSITGLGPILALNEDEALDYILSKNSSDIRKVTKSNLDENDKNETIESKNADVICDEMATNDGSLPLPIQYPSTSQDAADELLASPTSVKSEDFEAHCEVVITGNSLIGKGWSADNNDDLNASIHSNSSHGSSMIKTPEIKSLSSLVDQTNPFAETFSNMPNLRDIIKDIHRELKFTPDDVARSNIKIDPVTPEDPALQGLDFEVVPSSMTESFTVPELQSLIQQLGVLSREQGLDHQNYQCKGCQDLLGSTISRAKVCAYTGEYYCSNCMDPNVFIIPARVIHNWDFKRYPVSKKAALFLLEFQHHPWIDMKKLNPKIYCGVSDMAHLQDLRIQLNFLRAYIFTCREPVIEDLQKRIWPREHLYEHVHLYTISDLAQIPNGSLVLQLEKVVSFARSHVLDCWLCSQKGFICEVCKDPKILYPFETAGTYRCDECSSVFHDRCLNVNIPCPKCKRKQNRTNDTSLVDAVNACFNKL
Summary
Uniprot
H9JDE5
A0A2A4JEK6
A0A194R7D0
A0A194PKE8
A0A0L7LNR8
A0A2H1V7J4
+ More
A0A1E1W900 A0A2J7PHC8 A0A067QY45 A0A2J7PHC4 D6W845 A0A1Y1JW71 A0A151IEY4 A0A1W4X890 U4U1J0 A0A1B0CYT5 N6T901 A0A0J7KQT1 K7J291 E2BUT6 E2ADQ9 A0A151WMA1 E9ISM6 A0A195EUC8 A0A088AGT9 A0A0M9ABE2 A0A2A3EA75 A0A154PSJ7 A0A195E5U8 A0A195B0W3 F4WPF9 A0A158NR07 C9QPB2 B4I7Z9 Q9W293 B5X548 B4QH20 W8BRZ6 A0A1W4UM98 B3NNC8 A0A0A1XKS3 A0A026WRG0 A0A3L8DWL3 B4P8A7 W8B0J1 B4KMV4 B4LNE4 A0A0Q9X929 A0A1I8MSZ2 B4MQP8 A0A1B6G787 B4J8J6 A0A1B6E684 A0A0M4EEK2 A0A0M4EM58 A0A310SAZ0 A0A023EZS9 A0A224XCQ3 A0A1J1HTZ9 A0A1B6C9V8 A0A1Z5KXZ6 A0A224Z2K4 A0A131YPP1 A0A293LUI4 T1HMT1 A0A087UPG0 A0A2P6KTW2 A0A147BQB2 A0A0N7ZBD9 A0A443SVY4 A0A1W4WXP1 A0A0L8GLN0 A0A0L8GMK0 A0A0L8GLY3 A0A0L8GLJ8 A0A1B6DF18 E0VRE2 A0A132ADF2
A0A1E1W900 A0A2J7PHC8 A0A067QY45 A0A2J7PHC4 D6W845 A0A1Y1JW71 A0A151IEY4 A0A1W4X890 U4U1J0 A0A1B0CYT5 N6T901 A0A0J7KQT1 K7J291 E2BUT6 E2ADQ9 A0A151WMA1 E9ISM6 A0A195EUC8 A0A088AGT9 A0A0M9ABE2 A0A2A3EA75 A0A154PSJ7 A0A195E5U8 A0A195B0W3 F4WPF9 A0A158NR07 C9QPB2 B4I7Z9 Q9W293 B5X548 B4QH20 W8BRZ6 A0A1W4UM98 B3NNC8 A0A0A1XKS3 A0A026WRG0 A0A3L8DWL3 B4P8A7 W8B0J1 B4KMV4 B4LNE4 A0A0Q9X929 A0A1I8MSZ2 B4MQP8 A0A1B6G787 B4J8J6 A0A1B6E684 A0A0M4EEK2 A0A0M4EM58 A0A310SAZ0 A0A023EZS9 A0A224XCQ3 A0A1J1HTZ9 A0A1B6C9V8 A0A1Z5KXZ6 A0A224Z2K4 A0A131YPP1 A0A293LUI4 T1HMT1 A0A087UPG0 A0A2P6KTW2 A0A147BQB2 A0A0N7ZBD9 A0A443SVY4 A0A1W4WXP1 A0A0L8GLN0 A0A0L8GMK0 A0A0L8GLY3 A0A0L8GLJ8 A0A1B6DF18 E0VRE2 A0A132ADF2
Pubmed
19121390
26354079
26227816
24845553
18362917
19820115
+ More
28004739 23537049 20075255 20798317 21282665 21719571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24495485 25830018 24508170 30249741 17550304 25315136 25474469 28528879 28797301 26830274 29652888 20566863 26555130
28004739 23537049 20075255 20798317 21282665 21719571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 24495485 25830018 24508170 30249741 17550304 25315136 25474469 28528879 28797301 26830274 29652888 20566863 26555130
EMBL
BABH01030073
NWSH01001662
PCG70497.1
KQ460615
KPJ13733.1
KQ459602
+ More
KPI93483.1 JTDY01000455 KOB77097.1 ODYU01001058 SOQ36769.1 GDQN01007610 JAT83444.1 NEVH01025141 PNF15729.1 KK852888 KDR14325.1 PNF15730.1 KQ971307 EFA11621.2 GEZM01102066 GEZM01102065 GEZM01102064 GEZM01102063 GEZM01102062 GEZM01102061 GEZM01102057 GEZM01102056 JAV52010.1 KQ977835 KYM99382.1 KB631792 ERL86183.1 AJVK01009338 APGK01047183 KB741077 ENN74188.1 LBMM01004165 KMQ92742.1 AAZX01007799 GL450746 EFN80559.1 GL438820 EFN68421.1 KQ982944 KYQ48999.1 GL765434 EFZ16359.1 KQ981965 KYN31853.1 KQ435692 KOX81087.1 KZ288323 PBC28109.1 KQ435144 KZC14895.1 KQ979608 KYN20461.1 KQ976691 KYM77930.1 GL888243 EGI64054.1 ADTU01023720 BT099994 ACX54878.1 CH480824 EDW56724.1 AE013599 AAF46799.1 BT046167 ACI46555.1 CM000362 CM002911 EDX08166.1 KMY95708.1 GAMC01014576 JAB91979.1 CH954179 EDV55552.1 GBXI01002358 JAD11934.1 KK107139 EZA57679.1 QOIP01000003 RLU24465.1 CM000158 EDW91147.1 GAMC01014578 JAB91977.1 CH933808 EDW09876.1 CH940648 EDW61096.1 KRG04955.1 CH963849 EDW74437.1 GECZ01011464 JAS58305.1 CH916367 EDW01263.1 GEDC01026580 GEDC01008741 GEDC01003870 JAS10718.1 JAS28557.1 JAS33428.1 CP012524 ALC40472.1 ALC42438.1 ALC42792.1 KQ761843 OAD56595.1 GBBI01004416 JAC14296.1 GFTR01007672 JAW08754.1 CVRI01000013 CRK89633.1 GEDC01027026 JAS10272.1 GFJQ02007236 JAV99733.1 GFPF01009176 MAA20322.1 GEDV01008147 JAP80410.1 GFWV01007622 MAA32352.1 ACPB03015125 KK120873 KFM79249.1 MWRG01005371 PRD29738.1 GEGO01002448 JAR92956.1 GDRN01085203 GDRN01085202 GDRN01085201 GDRN01085200 JAI61376.1 NCKV01000090 RWS31676.1 KQ421289 KOF77853.1 KOF77855.1 KOF77854.1 KOF77852.1 GEDC01013005 JAS24293.1 DS235465 EEB15947.1 JXLN01012453 KPM08480.1
KPI93483.1 JTDY01000455 KOB77097.1 ODYU01001058 SOQ36769.1 GDQN01007610 JAT83444.1 NEVH01025141 PNF15729.1 KK852888 KDR14325.1 PNF15730.1 KQ971307 EFA11621.2 GEZM01102066 GEZM01102065 GEZM01102064 GEZM01102063 GEZM01102062 GEZM01102061 GEZM01102057 GEZM01102056 JAV52010.1 KQ977835 KYM99382.1 KB631792 ERL86183.1 AJVK01009338 APGK01047183 KB741077 ENN74188.1 LBMM01004165 KMQ92742.1 AAZX01007799 GL450746 EFN80559.1 GL438820 EFN68421.1 KQ982944 KYQ48999.1 GL765434 EFZ16359.1 KQ981965 KYN31853.1 KQ435692 KOX81087.1 KZ288323 PBC28109.1 KQ435144 KZC14895.1 KQ979608 KYN20461.1 KQ976691 KYM77930.1 GL888243 EGI64054.1 ADTU01023720 BT099994 ACX54878.1 CH480824 EDW56724.1 AE013599 AAF46799.1 BT046167 ACI46555.1 CM000362 CM002911 EDX08166.1 KMY95708.1 GAMC01014576 JAB91979.1 CH954179 EDV55552.1 GBXI01002358 JAD11934.1 KK107139 EZA57679.1 QOIP01000003 RLU24465.1 CM000158 EDW91147.1 GAMC01014578 JAB91977.1 CH933808 EDW09876.1 CH940648 EDW61096.1 KRG04955.1 CH963849 EDW74437.1 GECZ01011464 JAS58305.1 CH916367 EDW01263.1 GEDC01026580 GEDC01008741 GEDC01003870 JAS10718.1 JAS28557.1 JAS33428.1 CP012524 ALC40472.1 ALC42438.1 ALC42792.1 KQ761843 OAD56595.1 GBBI01004416 JAC14296.1 GFTR01007672 JAW08754.1 CVRI01000013 CRK89633.1 GEDC01027026 JAS10272.1 GFJQ02007236 JAV99733.1 GFPF01009176 MAA20322.1 GEDV01008147 JAP80410.1 GFWV01007622 MAA32352.1 ACPB03015125 KK120873 KFM79249.1 MWRG01005371 PRD29738.1 GEGO01002448 JAR92956.1 GDRN01085203 GDRN01085202 GDRN01085201 GDRN01085200 JAI61376.1 NCKV01000090 RWS31676.1 KQ421289 KOF77853.1 KOF77855.1 KOF77854.1 KOF77852.1 GEDC01013005 JAS24293.1 DS235465 EEB15947.1 JXLN01012453 KPM08480.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000235965
+ More
UP000027135 UP000007266 UP000078542 UP000192223 UP000030742 UP000092462 UP000019118 UP000036403 UP000002358 UP000008237 UP000000311 UP000075809 UP000078541 UP000005203 UP000053105 UP000242457 UP000076502 UP000078492 UP000078540 UP000007755 UP000005205 UP000001292 UP000000803 UP000000304 UP000192221 UP000008711 UP000053097 UP000279307 UP000002282 UP000009192 UP000008792 UP000095301 UP000007798 UP000001070 UP000092553 UP000183832 UP000015103 UP000054359 UP000288716 UP000053454 UP000009046
UP000027135 UP000007266 UP000078542 UP000192223 UP000030742 UP000092462 UP000019118 UP000036403 UP000002358 UP000008237 UP000000311 UP000075809 UP000078541 UP000005203 UP000053105 UP000242457 UP000076502 UP000078492 UP000078540 UP000007755 UP000005205 UP000001292 UP000000803 UP000000304 UP000192221 UP000008711 UP000053097 UP000279307 UP000002282 UP000009192 UP000008792 UP000095301 UP000007798 UP000001070 UP000092553 UP000183832 UP000015103 UP000054359 UP000288716 UP000053454 UP000009046
Interpro
ProteinModelPortal
H9JDE5
A0A2A4JEK6
A0A194R7D0
A0A194PKE8
A0A0L7LNR8
A0A2H1V7J4
+ More
A0A1E1W900 A0A2J7PHC8 A0A067QY45 A0A2J7PHC4 D6W845 A0A1Y1JW71 A0A151IEY4 A0A1W4X890 U4U1J0 A0A1B0CYT5 N6T901 A0A0J7KQT1 K7J291 E2BUT6 E2ADQ9 A0A151WMA1 E9ISM6 A0A195EUC8 A0A088AGT9 A0A0M9ABE2 A0A2A3EA75 A0A154PSJ7 A0A195E5U8 A0A195B0W3 F4WPF9 A0A158NR07 C9QPB2 B4I7Z9 Q9W293 B5X548 B4QH20 W8BRZ6 A0A1W4UM98 B3NNC8 A0A0A1XKS3 A0A026WRG0 A0A3L8DWL3 B4P8A7 W8B0J1 B4KMV4 B4LNE4 A0A0Q9X929 A0A1I8MSZ2 B4MQP8 A0A1B6G787 B4J8J6 A0A1B6E684 A0A0M4EEK2 A0A0M4EM58 A0A310SAZ0 A0A023EZS9 A0A224XCQ3 A0A1J1HTZ9 A0A1B6C9V8 A0A1Z5KXZ6 A0A224Z2K4 A0A131YPP1 A0A293LUI4 T1HMT1 A0A087UPG0 A0A2P6KTW2 A0A147BQB2 A0A0N7ZBD9 A0A443SVY4 A0A1W4WXP1 A0A0L8GLN0 A0A0L8GMK0 A0A0L8GLY3 A0A0L8GLJ8 A0A1B6DF18 E0VRE2 A0A132ADF2
A0A1E1W900 A0A2J7PHC8 A0A067QY45 A0A2J7PHC4 D6W845 A0A1Y1JW71 A0A151IEY4 A0A1W4X890 U4U1J0 A0A1B0CYT5 N6T901 A0A0J7KQT1 K7J291 E2BUT6 E2ADQ9 A0A151WMA1 E9ISM6 A0A195EUC8 A0A088AGT9 A0A0M9ABE2 A0A2A3EA75 A0A154PSJ7 A0A195E5U8 A0A195B0W3 F4WPF9 A0A158NR07 C9QPB2 B4I7Z9 Q9W293 B5X548 B4QH20 W8BRZ6 A0A1W4UM98 B3NNC8 A0A0A1XKS3 A0A026WRG0 A0A3L8DWL3 B4P8A7 W8B0J1 B4KMV4 B4LNE4 A0A0Q9X929 A0A1I8MSZ2 B4MQP8 A0A1B6G787 B4J8J6 A0A1B6E684 A0A0M4EEK2 A0A0M4EM58 A0A310SAZ0 A0A023EZS9 A0A224XCQ3 A0A1J1HTZ9 A0A1B6C9V8 A0A1Z5KXZ6 A0A224Z2K4 A0A131YPP1 A0A293LUI4 T1HMT1 A0A087UPG0 A0A2P6KTW2 A0A147BQB2 A0A0N7ZBD9 A0A443SVY4 A0A1W4WXP1 A0A0L8GLN0 A0A0L8GMK0 A0A0L8GLY3 A0A0L8GLJ8 A0A1B6DF18 E0VRE2 A0A132ADF2
PDB
2DWK
E-value=1.11587e-05,
Score=120
Ontologies
GO
Topology
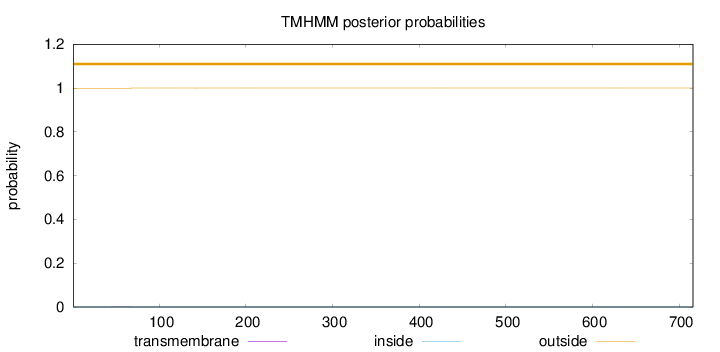
Length:
716
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01175
Exp number, first 60 AAs:
0.00615
Total prob of N-in:
0.00055
outside
1 - 716
Population Genetic Test Statistics
Pi
231.140516
Theta
199.149866
Tajima's D
0.546047
CLR
0.151696
CSRT
0.528323583820809
Interpretation
Uncertain
