Gene
KWMTBOMO09129
Pre Gene Modal
BGIBMGA007540
Annotation
PREDICTED:_GTPase-activating_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.95
Sequence
CDS
ATGAAAGAGATAAACAGTTATGGATTTAACATGGCGGACGATACGCGTAAAGTGCGAATCGAGGAAAGATTGAAAATAAAGATTGGTGAAGCAAAGAACTTGCCAGGTCGCAATCATGGTGCGGCGTGTCATCGAGACATATACTGCGTGCTGTCGCTCGATCAGGAGGAAATATTTCGTACGTCCACCATGGAGAGAACACTTAACCCATTTTTCGGAGAAGAGTTCCAGTTTGAAGTTCCCAGACGATTTCGGTACTTATCAATATACGTGTTCGATCGTGACAAACATCTCAAACAGGACAAAGTGCTCGGAAAAGTTGCAATCAAACGAGAAGACCTGCACCGATATAACAACACAGACCACTGGTTCGCGCTGCGACCGGTCGACGCAGACTCAGAGGTGCAGGGGAAAGCTTACATCGAACTCACCCTCGAGGATTCGAGAAAGCGGTTACGGATGGAGCCGAAGCCCCCCGACCCGGACCCGGCCGCCGACAACTGCTCGACGAAGAGCAAACAGAACAATCTCGTGAGATTATCGGAGTATTGTAAGGAGAGCATGAGATTAGGATCTTGCTCGAGCTCGACGAGCAAGAAGCTGCTGAGCGTGGCGACAGAGCCCGCCATGTCTCTGTCGCGGTCGGAGAGCCTGAAGGATCGAGCACAGCTGACGTCACGCTCGAAGCCGCCCCTGCACGCGATGCCAGAGACCGACACCTCGCCCACTCCCACCGCCGCCGACTACTGGTCCGACCCGGAGCGGCACTGCGCGGCCAGGGTCGGCTACGACACCATACGCCTGCGAGTGCTCGAATGCTCCGAGCTCACCACCAAGAATGGACAGTGCGACCCCTTCGCTCTCGTCACGTTACTCTACTCGAACGGACGGAGAGTCGAGAAACGCACCAAACCCAAAAAGAAGACTGTCAACCCTCAATTTGATGAAATATTTTGTTTTGATCTCGTAATGGAAGCGGACCCAAAAGACAAAGACGGCTCCCAAACGGCCGGCTTAGCGTGCGAAGCTAGAGTGTCGGTGTGGCACGATGCGACCACGCCCATATTTCTTGGCGAAGTGCGACTTCAGCTGCGGCAACCGACGCCCTCGCCCTTGCGAGCTTGGTTCTTCCTGACGTCGCGCGCGGGCGGCGCCCTGTCCCGCGGCGCCACCCCGCCCGGCACGCGGCTGTCGGCGGCGGGCAGCGCCACGCTGGGCTCGCTGAGGCTGCTGCTGCAGTACACCGTCGACCACGTCTTCCCCAAGAACCACTACCGCAGCCTCGAGGAACTCTTCCTCCGAAGTGTGCATCAGAAGCCCACGACAGCGTCGGCGGTGTACATCCTGGGCGAGATTGTTGCGAGCAAGACGGACGCAGCCCAACCCCTAGTAAGACTGTTTATGCATCACGATCTCATCGTTCCCATCGTCAAAGAACTCGCCGATTCCGAAATATCGGCTCTTACTGATGCGACCACAATATTCAGAGGCAACACACTGGTGTCTAAAATGATGGACGAGGCGATGCGGCTCAGCGGCGCGTCTTACTTACGTGCCGTGCTGCGGCCCACTCTCGCTGCTGTGCTCGCAGAGAAGCGACCCTGCGAGATAGACCCGGCCAGGGTCAAACCCTCTGCCGCTATTACCGCTAATTTGACCAACTTGAAAGACTACGTTGAACGGGTGTTCGGTGCGATAACGTCGTCGTACGCGATGTGCCCGGCCACGATGTGCAGACTGTTCGACGCGCTGCGGCAATGTGCGGCGCGACACTTCCCCAGGAACGCGGAAGTGCGGTATTCTGTCGTCTCGGGATTCATTTTCTTGAGATTCTTTGCGCCCGCCATACTCGGACCCAGGCTGTTTGACCTCACAACAGAACAAATTGATCCGCAAACCAACAGAACCTTGACATTGATCTCTAAGACGATCCAGTCGCTGGGGAACTTGGTGAGCAGTAGGTCTGCGCAGCAGGTCTGTAAGGAGGAGTACATGGCTGAACTGTACAGAAGCTTCTGCACGCCAAGACATGTGCAAGCAATAAAGCAGTTTCTAGAAATAATATCAACGAGCACAGACACACAGAACAACAACGTACAAGAGACGCCTGTATTACTTAAAGAAGGGACGATGATCAAACGTTCTGAAGGAGTGCGGGGCGGCGGCTCGCCCCGACGTCGCTGGACTCGCGCTCCGCACGCGCACGCCAAGCGGAGGCACTTCCGACTCACGAGCGGCGAGCTGGTGTGGTCCCGGTGCGGGGCGCGCGGCGGCGCCCAGCGACTGTCGCTGTCGGGCGTGCTCGCCGTGGCGCCCGTGGACTGTCCGCCGGCCGGCGCGCCGCTCGGGACCAACAACATATTTCAGATAGTGCACCACGAGCGCGTGATGTTTGTCCAGGCGAGTAACTGCGTGGAGCGAGCCGAGTGGGTGCGCATGTTGCTGGCGCTGTGCCGCCGCTCGCCCGCCGCGCCGCTGCACACCGGCTACTACGCAGACACGCACTGGACCTGTTGCGGTCGCACGGAAGCAGTAGCGGAAGGTTGCGGTGGGACAGAGAACGGCATGGAGGCGGGGGACGGCACCGCGGCTGGCGTCGCTCTGGCGCTGCGGCTCGACCCGGCGCGGGACCTGCAGCGGCTGCACGCGCTGCTCGTCTCGCACGCCGCCGCACTCGACGACAGGCTACACCGACCTCAACTAACTACGAGTATAGAAGAACGTGAGGCAGAAGCGGCTACCCTACGAGAGTTGCAAGAGGCTATGTTTAACCTAGAGCACCAGCATCGAATATACCGACGGTCGCTAGCTCGAGAAACTAAATACGGAAGCAAACAAGCGCCAATAGGTGACGACAACTATTTACACTTAGCCGGAGCAGGAGGGGTGGACTGCGGCGGGTTGCCGTGGCGAAATTGGCGCAGCGACGCTGCGGTCGATCTCGACGGTAAGCCGCTGCGGCGCAACGACATGACTTCGAGCACAGAGTCGCTGCGCCGGCACGCCGACGAGCGCTCGTCCGGTAAGTCGACGCGCTCCGCCGGCAGCGGGTACCTCGCCATGTCCTGCTTGCGGCACGAGCCGGCCGACGACAGCGACGCGTCGGAGCGACCCGCTCGGCCGCCCAAACCGGCACGACTATCGCCTCCCCGCGCCTTCATCGACCCTCCTGCCGACTATGTGCCGGTTGAACCTCCGCCGCGATGTCGAGAGTACGAGCTGATGTCAGACTTCGTGAGTCGGGCCCGCGCTCTTGATGATGAGCCTCCTCCCGCGGTGCCGCCCCGCACCCGTCCCCGCCCCGTCGAGCCGCCGCGTCCCTCTAGCGCTGCCGACGCCGTCTCCACAGATGACAGTTTGATCGATGAACTACAACGCGGATCATACTGTGTGCTCAGAGTGTGCGAAGACGAATCTCCAGTACCGGAGAACTTTGGGGAGGAGGCTTCGGAGGCCGTCGACCAATCTAATGAACCGTTCCAACGTTCGAGCGTACAGCGGAAATCAAACCCGACCCGATCTCGGATCAGTCCGACTCGACCCCTCAAAACATCCAGTTCGACTTCTAACGTGCCGGAAGCAGCTCGCACCGCTTCCTCATCGCGGCTTTTCGCTGAACGTCTAACTCCCTTTTTTTTTAAACGGAAACAAAAACCTCTTGAAGAGGCGACGGGCAACTCCCGACGTGAAGAAAAAGACAACATGTTCGGGCGCTTGACTCCGCGTTTCGGTCAAGCGAGGCTGACGGGGCGCGGACGGTCGCTGGAGCTGACTGCGGAGGAGGCTCGTCCGCGGCGCATCGAGCGCTCCGCCACCACAGTCAACATCCCCCCGCGCCCCATGAACTCATCGATCGTCGCCAACTTGAAGTTCCTGAGTCTGCACAGGTACGGGCCGCAGGAGGCGAACCGAAAGTCCCCAGGCGAGGCGGAAGGTGCGTCGTGGCGGCCGCAGCATTCCGCCTCGCTGTTGGCCACAGCCTCTACCGACATCCGCCTTTAA
Protein
MKEINSYGFNMADDTRKVRIEERLKIKIGEAKNLPGRNHGAACHRDIYCVLSLDQEEIFRTSTMERTLNPFFGEEFQFEVPRRFRYLSIYVFDRDKHLKQDKVLGKVAIKREDLHRYNNTDHWFALRPVDADSEVQGKAYIELTLEDSRKRLRMEPKPPDPDPAADNCSTKSKQNNLVRLSEYCKESMRLGSCSSSTSKKLLSVATEPAMSLSRSESLKDRAQLTSRSKPPLHAMPETDTSPTPTAADYWSDPERHCAARVGYDTIRLRVLECSELTTKNGQCDPFALVTLLYSNGRRVEKRTKPKKKTVNPQFDEIFCFDLVMEADPKDKDGSQTAGLACEARVSVWHDATTPIFLGEVRLQLRQPTPSPLRAWFFLTSRAGGALSRGATPPGTRLSAAGSATLGSLRLLLQYTVDHVFPKNHYRSLEELFLRSVHQKPTTASAVYILGEIVASKTDAAQPLVRLFMHHDLIVPIVKELADSEISALTDATTIFRGNTLVSKMMDEAMRLSGASYLRAVLRPTLAAVLAEKRPCEIDPARVKPSAAITANLTNLKDYVERVFGAITSSYAMCPATMCRLFDALRQCAARHFPRNAEVRYSVVSGFIFLRFFAPAILGPRLFDLTTEQIDPQTNRTLTLISKTIQSLGNLVSSRSAQQVCKEEYMAELYRSFCTPRHVQAIKQFLEIISTSTDTQNNNVQETPVLLKEGTMIKRSEGVRGGGSPRRRWTRAPHAHAKRRHFRLTSGELVWSRCGARGGAQRLSLSGVLAVAPVDCPPAGAPLGTNNIFQIVHHERVMFVQASNCVERAEWVRMLLALCRRSPAAPLHTGYYADTHWTCCGRTEAVAEGCGGTENGMEAGDGTAAGVALALRLDPARDLQRLHALLVSHAAALDDRLHRPQLTTSIEEREAEAATLRELQEAMFNLEHQHRIYRRSLARETKYGSKQAPIGDDNYLHLAGAGGVDCGGLPWRNWRSDAAVDLDGKPLRRNDMTSSTESLRRHADERSSGKSTRSAGSGYLAMSCLRHEPADDSDASERPARPPKPARLSPPRAFIDPPADYVPVEPPPRCREYELMSDFVSRARALDDEPPPAVPPRTRPRPVEPPRPSSAADAVSTDDSLIDELQRGSYCVLRVCEDESPVPENFGEEASEAVDQSNEPFQRSSVQRKSNPTRSRISPTRPLKTSSSTSNVPEAARTASSSRLFAERLTPFFFKRKQKPLEEATGNSRREEKDNMFGRLTPRFGQARLTGRGRSLELTAEEARPRRIERSATTVNIPPRPMNSSIVANLKFLSLHRYGPQEANRKSPGEAEGASWRPQHSASLLATASTDIRL
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF48350
SSF48350
Gene 3D
ProteinModelPortal
PDB
1WQ1
E-value=2.74186e-32,
Score=352
Ontologies
GO
PANTHER
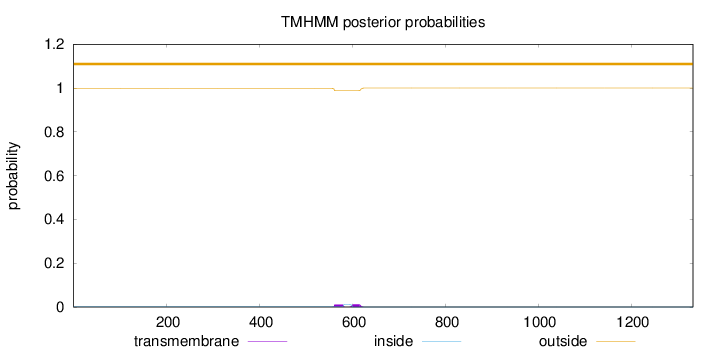
Topology
Length:
1333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.490410000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00090
outside
1 - 1333
Population Genetic Test Statistics
Pi
19.447571
Theta
163.09301
Tajima's D
0.469291
CLR
0.671651
CSRT
0.509624518774061
Interpretation
Uncertain
