Gene
KWMTBOMO09127 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007559
Annotation
PREDICTED:_transmembrane_protein_256_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.043
Sequence
CDS
ATGATAAGAGCTCCATTGTGGCAACTTGCTCAAGAGGCCGGTCCTTTTGTTAGGCTGGCAGGACTAAGTGGTGCTGCTGCTGTAATCCTTGGTGCTATGGGAGCACATCGAACTTTCCCTGAGACTGAGGCTAAAGACGACTTGAAGAAAATATTTGAAACTGCCAATAGATTCCATTTTCTACATACCCTCGCTTTAGTAACCGTTCCTCTGTGTAGAAGACCGTATATAGCTGGTGCATTTTTTATTGCTGGTATGGGAATGTTCTGTGGAACTTGTTACTACCATGCTTTCACTGGTGATAGAAAATTGCGTCAAGTGACACCTCTCGGTGGGACCTGCTTAATCTTAGGATGGATTGCAATGGTTGGATGTGAATATTGGTTGGATGCTTCAATACTGTTGTTCATGGTGTGTATACCTTTGTAA
Protein
MIRAPLWQLAQEAGPFVRLAGLSGAAAVILGAMGAHRTFPETEAKDDLKKIFETANRFHFLHTLALVTVPLCRRPYIAGAFFIAGMGMFCGTCYYHAFTGDRKLRQVTPLGGTCLILGWIAMVGCEYWLDASILLFMVCIPL
Summary
Uniprot
H9JDG2
A0A3S2P7W3
A0A0L7L762
S4Q099
A0A212FCE8
A0A2A4JZP0
+ More
A0A2H1V8H9 I4DL41 A0A194PQS2 I4DMX2 A0A194RCU1 A0A0L7QM55 A0A232F5S8 D6WKR5 K7IVT7 A0A224XZ65 A0A170Y2S0 A0A0V0G7A5 A0A069DPQ6 A0A023F8M8 A0A2R7WK27 A0A0N7Z8K7 R4FPR1 A0A0A9WV31 A0A1W4X3D0 A0A0A9Z1P3 N6TM80 J3JTL9 A0A151J6C3 A0A1B6ISR2 A0A1B6IEM7 E2B562 A0A1B6HMZ0 A0A1B6G254 A0A1B6H7A1 E2AG79 A0A1B6MLY9 A0A1B6F9X0 A0A195F917 A0A1B6G535 A0A1B6FEG1 A0A1B6MKH7 A0A195CFQ2 A0A1B6LU26 A0A1B6M772 A0A151WFM6 A0A1B6CVM9 A0A1B6C213 A0A1B6D1E7 A0A195BY34 A0A158NCA8 F4X836 R4V263 A0A067RKL5 A0A0M9A5R0 A0A2P8ZD02 A0A2P2HYC2 A0A0A1WWB4 A0A423T7S1 A0A0T6AUL6 A0A1L8D9J5 A0A034WLI9 A0A1L8D9X7 A0A0C9R7P0 A0A0K8V366 W8C187 A0A0K8TPY6 A0A444TB61 A0A0P4WI40 E2BD41 A0A087ZTD8 A0A2A3EBH6 A0A154PHG6 A0A182VNA0 A0A0P5P7X1 A0A023FR98 A0A182X9K3 A0A0P5WPF0 A0A0N8B1X4 A0A1C7CZS6 A0NH70 A0A182HKI6 A0A0P5M2U3 A0A023FPK5 A0A444SSB2 A0A0P5CS23 A0A0P5UPY5 A0A0P5CV16 E9G2A3 A0A0P5BY04 A0A0P6AI33 A0A182T9D0 A0A0P6IF34 A0A0P5TYE5 A0A182YBE4 A0A1I8JSV2 A0A0P6D6I5 U5ET33 A0A2M3ZIW1 A0A2M3ZAV0
A0A2H1V8H9 I4DL41 A0A194PQS2 I4DMX2 A0A194RCU1 A0A0L7QM55 A0A232F5S8 D6WKR5 K7IVT7 A0A224XZ65 A0A170Y2S0 A0A0V0G7A5 A0A069DPQ6 A0A023F8M8 A0A2R7WK27 A0A0N7Z8K7 R4FPR1 A0A0A9WV31 A0A1W4X3D0 A0A0A9Z1P3 N6TM80 J3JTL9 A0A151J6C3 A0A1B6ISR2 A0A1B6IEM7 E2B562 A0A1B6HMZ0 A0A1B6G254 A0A1B6H7A1 E2AG79 A0A1B6MLY9 A0A1B6F9X0 A0A195F917 A0A1B6G535 A0A1B6FEG1 A0A1B6MKH7 A0A195CFQ2 A0A1B6LU26 A0A1B6M772 A0A151WFM6 A0A1B6CVM9 A0A1B6C213 A0A1B6D1E7 A0A195BY34 A0A158NCA8 F4X836 R4V263 A0A067RKL5 A0A0M9A5R0 A0A2P8ZD02 A0A2P2HYC2 A0A0A1WWB4 A0A423T7S1 A0A0T6AUL6 A0A1L8D9J5 A0A034WLI9 A0A1L8D9X7 A0A0C9R7P0 A0A0K8V366 W8C187 A0A0K8TPY6 A0A444TB61 A0A0P4WI40 E2BD41 A0A087ZTD8 A0A2A3EBH6 A0A154PHG6 A0A182VNA0 A0A0P5P7X1 A0A023FR98 A0A182X9K3 A0A0P5WPF0 A0A0N8B1X4 A0A1C7CZS6 A0NH70 A0A182HKI6 A0A0P5M2U3 A0A023FPK5 A0A444SSB2 A0A0P5CS23 A0A0P5UPY5 A0A0P5CV16 E9G2A3 A0A0P5BY04 A0A0P6AI33 A0A182T9D0 A0A0P6IF34 A0A0P5TYE5 A0A182YBE4 A0A1I8JSV2 A0A0P6D6I5 U5ET33 A0A2M3ZIW1 A0A2M3ZAV0
Pubmed
EMBL
BABH01030078
RSAL01000223
RVE43997.1
JTDY01002476
KOB71337.1
GAIX01000158
+ More
JAA92402.1 AGBW02009204 OWR51387.1 NWSH01000352 PCG77114.1 ODYU01001228 SOQ37153.1 AK402009 BAM18631.1 KQ459602 KPI93480.1 AK402640 BAM19262.1 KQ460615 KPJ13726.1 KQ414894 KOC59697.1 NNAY01000939 OXU25823.1 KQ971342 EFA03033.1 GFTR01002516 JAW13910.1 GEMB01003726 JAR99494.1 GECL01002909 JAP03215.1 GBGD01003089 JAC85800.1 GBBI01000995 JAC17717.1 KK854966 PTY19997.1 GDKW01003075 JAI53520.1 ACPB03005605 GAHY01000749 JAA76761.1 GBHO01031267 GDHC01011618 JAG12337.1 JAQ07011.1 GBHO01005215 GBRD01013588 GDHC01018426 JAG38389.1 JAG52238.1 JAQ00203.1 APGK01018912 KB740085 KB632303 ENN81559.1 ERL91824.1 BT126576 AEE61540.1 KQ979863 KYN18755.1 GECU01017737 JAS89969.1 GECU01022310 JAS85396.1 GL445725 EFN89156.1 GECU01031659 JAS76047.1 GECZ01013254 JAS56515.1 GECU01037099 JAS70607.1 GL439266 EFN67570.1 GEBQ01003023 JAT36954.1 GECZ01022759 JAS47010.1 KQ981727 KYN37100.1 GECZ01012232 JAS57537.1 GECZ01021184 JAS48585.1 GEBQ01003618 JAT36359.1 KQ977873 KYM99131.1 GEBQ01012807 JAT27170.1 GEBQ01008195 JAT31782.1 KQ983211 KYQ46640.1 GEDC01019742 JAS17556.1 GEDC01029747 JAS07551.1 GEDC01017822 JAS19476.1 KQ976396 KYM92853.1 ADTU01011778 ADTU01011779 GL888912 EGI57385.1 KC740993 AGM32817.1 KK852413 KDR24416.1 KQ435736 KOX77016.1 PYGN01000097 PSN54374.1 IACF01001047 LAB66774.1 GBXI01011569 JAD02723.1 QCYY01002142 ROT72494.1 LJIG01022814 KRT78581.1 GFDF01011057 JAV03027.1 GAKP01002511 GAKP01002510 JAC56441.1 GFDF01010954 JAV03130.1 GBYB01008842 JAG78609.1 GDHF01018965 JAI33349.1 GAMC01003647 GAMC01003646 JAC02909.1 GDAI01001141 JAI16462.1 SAUD01005397 RXG67213.1 GDRN01049333 GDRN01049332 JAI66572.1 GL447563 EFN86383.1 KZ288292 PBC29087.1 KQ434905 KZC11247.1 GDIQ01136700 JAL15026.1 GBBK01001244 JAC23238.1 GDIP01096118 JAM07597.1 GDIQ01220867 JAK30858.1 AAAB01008987 EAU75720.1 APCN01000831 GDIQ01161393 JAK90332.1 GBBK01001243 JAC23239.1 SAUD01024938 RXG56285.1 GDIP01167655 JAJ55747.1 GDIP01127414 JAL76300.1 GDIP01169302 JAJ54100.1 GL732530 EFX86338.1 GDIP01178349 JAJ45053.1 GDIP01029464 JAM74251.1 GDIQ01022490 JAN72247.1 GDIP01122612 JAL81102.1 GDIQ01089968 JAN04769.1 GANO01002112 JAB57759.1 GGFM01007720 MBW28471.1 GGFM01004915 MBW25666.1
JAA92402.1 AGBW02009204 OWR51387.1 NWSH01000352 PCG77114.1 ODYU01001228 SOQ37153.1 AK402009 BAM18631.1 KQ459602 KPI93480.1 AK402640 BAM19262.1 KQ460615 KPJ13726.1 KQ414894 KOC59697.1 NNAY01000939 OXU25823.1 KQ971342 EFA03033.1 GFTR01002516 JAW13910.1 GEMB01003726 JAR99494.1 GECL01002909 JAP03215.1 GBGD01003089 JAC85800.1 GBBI01000995 JAC17717.1 KK854966 PTY19997.1 GDKW01003075 JAI53520.1 ACPB03005605 GAHY01000749 JAA76761.1 GBHO01031267 GDHC01011618 JAG12337.1 JAQ07011.1 GBHO01005215 GBRD01013588 GDHC01018426 JAG38389.1 JAG52238.1 JAQ00203.1 APGK01018912 KB740085 KB632303 ENN81559.1 ERL91824.1 BT126576 AEE61540.1 KQ979863 KYN18755.1 GECU01017737 JAS89969.1 GECU01022310 JAS85396.1 GL445725 EFN89156.1 GECU01031659 JAS76047.1 GECZ01013254 JAS56515.1 GECU01037099 JAS70607.1 GL439266 EFN67570.1 GEBQ01003023 JAT36954.1 GECZ01022759 JAS47010.1 KQ981727 KYN37100.1 GECZ01012232 JAS57537.1 GECZ01021184 JAS48585.1 GEBQ01003618 JAT36359.1 KQ977873 KYM99131.1 GEBQ01012807 JAT27170.1 GEBQ01008195 JAT31782.1 KQ983211 KYQ46640.1 GEDC01019742 JAS17556.1 GEDC01029747 JAS07551.1 GEDC01017822 JAS19476.1 KQ976396 KYM92853.1 ADTU01011778 ADTU01011779 GL888912 EGI57385.1 KC740993 AGM32817.1 KK852413 KDR24416.1 KQ435736 KOX77016.1 PYGN01000097 PSN54374.1 IACF01001047 LAB66774.1 GBXI01011569 JAD02723.1 QCYY01002142 ROT72494.1 LJIG01022814 KRT78581.1 GFDF01011057 JAV03027.1 GAKP01002511 GAKP01002510 JAC56441.1 GFDF01010954 JAV03130.1 GBYB01008842 JAG78609.1 GDHF01018965 JAI33349.1 GAMC01003647 GAMC01003646 JAC02909.1 GDAI01001141 JAI16462.1 SAUD01005397 RXG67213.1 GDRN01049333 GDRN01049332 JAI66572.1 GL447563 EFN86383.1 KZ288292 PBC29087.1 KQ434905 KZC11247.1 GDIQ01136700 JAL15026.1 GBBK01001244 JAC23238.1 GDIP01096118 JAM07597.1 GDIQ01220867 JAK30858.1 AAAB01008987 EAU75720.1 APCN01000831 GDIQ01161393 JAK90332.1 GBBK01001243 JAC23239.1 SAUD01024938 RXG56285.1 GDIP01167655 JAJ55747.1 GDIP01127414 JAL76300.1 GDIP01169302 JAJ54100.1 GL732530 EFX86338.1 GDIP01178349 JAJ45053.1 GDIP01029464 JAM74251.1 GDIQ01022490 JAN72247.1 GDIP01122612 JAL81102.1 GDIQ01089968 JAN04769.1 GANO01002112 JAB57759.1 GGFM01007720 MBW28471.1 GGFM01004915 MBW25666.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000007151
UP000218220
UP000053268
+ More
UP000053240 UP000053825 UP000215335 UP000007266 UP000002358 UP000015103 UP000192223 UP000019118 UP000030742 UP000078492 UP000008237 UP000000311 UP000078541 UP000078542 UP000075809 UP000078540 UP000005205 UP000007755 UP000027135 UP000053105 UP000245037 UP000283509 UP000288706 UP000005203 UP000242457 UP000076502 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000000305 UP000075901 UP000076408 UP000069272
UP000053240 UP000053825 UP000215335 UP000007266 UP000002358 UP000015103 UP000192223 UP000019118 UP000030742 UP000078492 UP000008237 UP000000311 UP000078541 UP000078542 UP000075809 UP000078540 UP000005205 UP000007755 UP000027135 UP000053105 UP000245037 UP000283509 UP000288706 UP000005203 UP000242457 UP000076502 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000000305 UP000075901 UP000076408 UP000069272
PRIDE
Pfam
Interpro
SUPFAM
SSF53300
SSF53300
ProteinModelPortal
H9JDG2
A0A3S2P7W3
A0A0L7L762
S4Q099
A0A212FCE8
A0A2A4JZP0
+ More
A0A2H1V8H9 I4DL41 A0A194PQS2 I4DMX2 A0A194RCU1 A0A0L7QM55 A0A232F5S8 D6WKR5 K7IVT7 A0A224XZ65 A0A170Y2S0 A0A0V0G7A5 A0A069DPQ6 A0A023F8M8 A0A2R7WK27 A0A0N7Z8K7 R4FPR1 A0A0A9WV31 A0A1W4X3D0 A0A0A9Z1P3 N6TM80 J3JTL9 A0A151J6C3 A0A1B6ISR2 A0A1B6IEM7 E2B562 A0A1B6HMZ0 A0A1B6G254 A0A1B6H7A1 E2AG79 A0A1B6MLY9 A0A1B6F9X0 A0A195F917 A0A1B6G535 A0A1B6FEG1 A0A1B6MKH7 A0A195CFQ2 A0A1B6LU26 A0A1B6M772 A0A151WFM6 A0A1B6CVM9 A0A1B6C213 A0A1B6D1E7 A0A195BY34 A0A158NCA8 F4X836 R4V263 A0A067RKL5 A0A0M9A5R0 A0A2P8ZD02 A0A2P2HYC2 A0A0A1WWB4 A0A423T7S1 A0A0T6AUL6 A0A1L8D9J5 A0A034WLI9 A0A1L8D9X7 A0A0C9R7P0 A0A0K8V366 W8C187 A0A0K8TPY6 A0A444TB61 A0A0P4WI40 E2BD41 A0A087ZTD8 A0A2A3EBH6 A0A154PHG6 A0A182VNA0 A0A0P5P7X1 A0A023FR98 A0A182X9K3 A0A0P5WPF0 A0A0N8B1X4 A0A1C7CZS6 A0NH70 A0A182HKI6 A0A0P5M2U3 A0A023FPK5 A0A444SSB2 A0A0P5CS23 A0A0P5UPY5 A0A0P5CV16 E9G2A3 A0A0P5BY04 A0A0P6AI33 A0A182T9D0 A0A0P6IF34 A0A0P5TYE5 A0A182YBE4 A0A1I8JSV2 A0A0P6D6I5 U5ET33 A0A2M3ZIW1 A0A2M3ZAV0
A0A2H1V8H9 I4DL41 A0A194PQS2 I4DMX2 A0A194RCU1 A0A0L7QM55 A0A232F5S8 D6WKR5 K7IVT7 A0A224XZ65 A0A170Y2S0 A0A0V0G7A5 A0A069DPQ6 A0A023F8M8 A0A2R7WK27 A0A0N7Z8K7 R4FPR1 A0A0A9WV31 A0A1W4X3D0 A0A0A9Z1P3 N6TM80 J3JTL9 A0A151J6C3 A0A1B6ISR2 A0A1B6IEM7 E2B562 A0A1B6HMZ0 A0A1B6G254 A0A1B6H7A1 E2AG79 A0A1B6MLY9 A0A1B6F9X0 A0A195F917 A0A1B6G535 A0A1B6FEG1 A0A1B6MKH7 A0A195CFQ2 A0A1B6LU26 A0A1B6M772 A0A151WFM6 A0A1B6CVM9 A0A1B6C213 A0A1B6D1E7 A0A195BY34 A0A158NCA8 F4X836 R4V263 A0A067RKL5 A0A0M9A5R0 A0A2P8ZD02 A0A2P2HYC2 A0A0A1WWB4 A0A423T7S1 A0A0T6AUL6 A0A1L8D9J5 A0A034WLI9 A0A1L8D9X7 A0A0C9R7P0 A0A0K8V366 W8C187 A0A0K8TPY6 A0A444TB61 A0A0P4WI40 E2BD41 A0A087ZTD8 A0A2A3EBH6 A0A154PHG6 A0A182VNA0 A0A0P5P7X1 A0A023FR98 A0A182X9K3 A0A0P5WPF0 A0A0N8B1X4 A0A1C7CZS6 A0NH70 A0A182HKI6 A0A0P5M2U3 A0A023FPK5 A0A444SSB2 A0A0P5CS23 A0A0P5UPY5 A0A0P5CV16 E9G2A3 A0A0P5BY04 A0A0P6AI33 A0A182T9D0 A0A0P6IF34 A0A0P5TYE5 A0A182YBE4 A0A1I8JSV2 A0A0P6D6I5 U5ET33 A0A2M3ZIW1 A0A2M3ZAV0
Ontologies
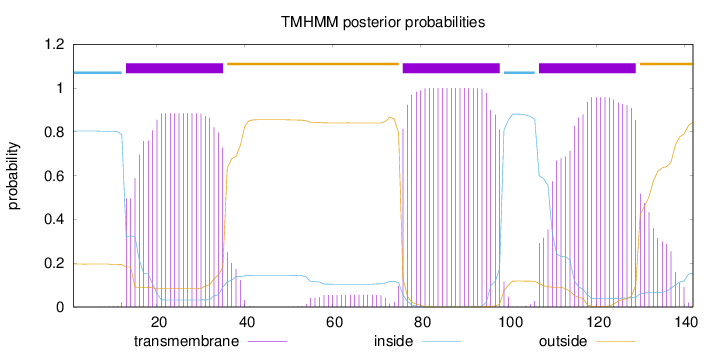
Topology
Length:
142
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
64.11617
Exp number, first 60 AAs:
19.51117
Total prob of N-in:
0.80395
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 75
TMhelix
76 - 98
inside
99 - 106
TMhelix
107 - 129
outside
130 - 142
Population Genetic Test Statistics
Pi
47.707572
Theta
180.285188
Tajima's D
-1.602423
CLR
5842.335493
CSRT
0.0467476626168692
Interpretation
Uncertain
