Gene
KWMTBOMO09117 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007566
Annotation
PREDICTED:_prenylated_Rab_acceptor_protein_1_isoform_X3_[Bombyx_mori]
Full name
PRA1 family protein
Location in the cell
PlasmaMembrane Reliability : 4.745
Sequence
CDS
ATGTCTGAAAATGTAAAGATTGATCTGTCAGGTGAAATAAACGCAACAACAGAAAAGAAAGGTTTCCAAAAGTTGTTGCAGCATGTACGTAGTGGTGCGGCTCCTGCATTGTTGATGGGAGTTTTATCTTCACGCAGGCCATGGACACAATTTGTTGCCACTGACAACTTTAAGGTACCAGTATCTATACCAAGATTGTCTCGAAGATTCTATCGTAACATAGAATATTTTCAAGCTAATTACCTAATTGTATTTCTGGGACTATTTGCCTATTGCCTCATAACATCACCACTGTTGCTGATCACTATGGTGGCCAGTTTCTTTGGATATAGAAAATTAACTTCAGGGCCGAATACTTGGAAGATTGGTGGTTGGGAGTTAACGAAAACACAGCAGTATGTTATAGCTGCTGCCGGATCCATGGCACTCTGTTGGCTTGCTGGAGCTGGAGCCGTTTTATTCTGGGTTTTAGGTGCAACGGTAACCGTAGTCGCCCTTCATGCAAGCTTCTTCGATGCAGAGGCACTACCTAATCTTGAAGACCCGGAGCAGTTTCCGATGATTGAGCAAGTGTGA
Protein
MSENVKIDLSGEINATTEKKGFQKLLQHVRSGAAPALLMGVLSSRRPWTQFVATDNFKVPVSIPRLSRRFYRNIEYFQANYLIVFLGLFAYCLITSPLLLITMVASFFGYRKLTSGPNTWKIGGWELTKTQQYVIAAAGSMALCWLAGAGAVLFWVLGATVTVVALHASFFDAEALPNLEDPEQFPMIEQV
Summary
Similarity
Belongs to the PRA1 family.
Uniprot
A0A2A4JJY5
A0A2A4JKY1
A0A2H1VMU1
A0A1E1WPF5
A0A194RCT2
A0A194PKD3
+ More
A0A437BGA1 S4P9K8 V5GF64 A0A1Y1K4D3 A0A2P8Z497 A0A067RIM0 A0A1W4XME8 A0A1A9WP35 A0A2J7Q588 A0A034WK12 A0A0K8TZ36 A0A1Q3FKD3 A0A182QGQ3 A0A182JIB3 D2A449 A0A1L8EE76 B0WR48 A0A182P401 A0A084VEN4 A0A182RBA1 A0A023EJD0 A0A182W111 A0A182URF4 A0A182U131 A0A182WVJ3 Q5TT69 Q1HQQ3 A0A182MF79 A0A1L8DUX5 A0A182I972 A0A182YG41 A0A182K0V2 A0A336L0L8 A0A1I8PIG9 A0A1B0D3Z2 A0A1B6CTJ3 A0A336ME42 A0A182NCH2 T1E285 T1PDJ0 A0A2M3ZKX3 A0A2M3Z2E7 A0A2M4C044 W5JG84 A0A2M4A4Z3 A0A182F9K9 A0A2M4C0Q4 A0A0L0CE71 A0A3R7MAW9 A0A1B0AKK1 A0A1B0A9A8 A0A1A9UIY1 D3TMZ2 A0A0M5IXW8 A0A1D2MZY3 E0VSW2 A0A1B0CPX4 B4MRI4 U5EJZ4 B4J9H2 B4KNT6 A0A131XXD3 B3MF38 B7PPW4 A0A0J9R986 B4HMA6 B4QHU3 A1Z823 A0A1B0CQN0 A0A0K8RI51 Q8I1H0 A0A1B6E3I8 B4LJX1 B4NX22 A0A131XFI4 A0A0P4WE99 A0A1W4UZ28 L7M347 A0A224YWC8 A0A131Z3S8 A0A0N0BGF1 A0A0C9SBA7 E6PBW4 A0A444TH27 G3MK76 A0A226D4N5 R4FM62 A0A2G8KWF2 A0A023G6B3 A0A3B0J4Q5 A0A0B8RQM5
A0A437BGA1 S4P9K8 V5GF64 A0A1Y1K4D3 A0A2P8Z497 A0A067RIM0 A0A1W4XME8 A0A1A9WP35 A0A2J7Q588 A0A034WK12 A0A0K8TZ36 A0A1Q3FKD3 A0A182QGQ3 A0A182JIB3 D2A449 A0A1L8EE76 B0WR48 A0A182P401 A0A084VEN4 A0A182RBA1 A0A023EJD0 A0A182W111 A0A182URF4 A0A182U131 A0A182WVJ3 Q5TT69 Q1HQQ3 A0A182MF79 A0A1L8DUX5 A0A182I972 A0A182YG41 A0A182K0V2 A0A336L0L8 A0A1I8PIG9 A0A1B0D3Z2 A0A1B6CTJ3 A0A336ME42 A0A182NCH2 T1E285 T1PDJ0 A0A2M3ZKX3 A0A2M3Z2E7 A0A2M4C044 W5JG84 A0A2M4A4Z3 A0A182F9K9 A0A2M4C0Q4 A0A0L0CE71 A0A3R7MAW9 A0A1B0AKK1 A0A1B0A9A8 A0A1A9UIY1 D3TMZ2 A0A0M5IXW8 A0A1D2MZY3 E0VSW2 A0A1B0CPX4 B4MRI4 U5EJZ4 B4J9H2 B4KNT6 A0A131XXD3 B3MF38 B7PPW4 A0A0J9R986 B4HMA6 B4QHU3 A1Z823 A0A1B0CQN0 A0A0K8RI51 Q8I1H0 A0A1B6E3I8 B4LJX1 B4NX22 A0A131XFI4 A0A0P4WE99 A0A1W4UZ28 L7M347 A0A224YWC8 A0A131Z3S8 A0A0N0BGF1 A0A0C9SBA7 E6PBW4 A0A444TH27 G3MK76 A0A226D4N5 R4FM62 A0A2G8KWF2 A0A023G6B3 A0A3B0J4Q5 A0A0B8RQM5
Pubmed
26354079
23622113
28004739
29403074
24845553
25348373
+ More
18362917 19820115 24438588 24945155 12364791 14747013 17210077 17204158 17510324 25244985 24330624 25315136 20920257 23761445 26108605 20353571 27289101 20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 28049606 25576852 28797301 26830274 26131772 22216098 29023486 25476704
18362917 19820115 24438588 24945155 12364791 14747013 17210077 17204158 17510324 25244985 24330624 25315136 20920257 23761445 26108605 20353571 27289101 20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 28049606 25576852 28797301 26830274 26131772 22216098 29023486 25476704
EMBL
NWSH01001151
PCG72377.1
PCG72378.1
ODYU01003413
SOQ42149.1
GDQN01003679
+ More
GDQN01002196 JAT87375.1 JAT88858.1 KQ460615 KPJ13716.1 KQ459602 KPI93468.1 RSAL01000065 RVE49439.1 GAIX01003714 JAA88846.1 GALX01005852 JAB62614.1 GEZM01093128 JAV56334.1 PYGN01000202 PSN51324.1 KK852658 KDR19145.1 NEVH01017591 PNF23740.1 GAKP01004482 JAC54470.1 GDHF01032600 JAI19714.1 GFDL01006985 JAV28060.1 AXCN02001187 KQ971348 EFA04845.1 GFDG01001787 JAV17012.1 DS232051 EDS33154.1 ATLV01012285 KE524778 KFB36428.1 GAPW01004664 JAC08934.1 AAAB01008888 EAL40611.2 DQ440391 CH477693 ABF18424.1 EAT37197.1 AXCM01000042 GFDF01003846 JAV10238.1 APCN01003212 UFQS01001411 UFQT01001411 SSX10746.1 SSX30428.1 AJVK01023821 GEDC01020643 JAS16655.1 UFQS01001048 UFQT01001048 SSX08700.1 SSX28614.1 GALA01001445 JAA93407.1 KA646200 AFP60829.1 GGFM01008430 MBW29181.1 GGFM01001935 MBW22686.1 GGFJ01009556 MBW58697.1 ADMH02001608 ETN61824.1 GGFK01002546 MBW35867.1 GGFJ01009744 MBW58885.1 JRES01000505 KNC30541.1 QCYY01001600 ROT76863.1 JXJN01025954 CCAG010011323 EZ422794 ADD19070.1 CP012524 ALC42144.1 LJIJ01000340 ODM98596.1 DS235758 EEB16468.1 AJWK01022757 CH963850 EDW74723.1 GANO01002011 JAB57860.1 CH916367 EDW01453.1 CH933808 EDW08981.1 GEFM01003882 JAP71914.1 CH902619 EDV37666.1 ABJB010403120 ABJB010413655 DS761162 EEC08636.1 CM002911 KMY92627.1 CH480816 EDW47183.1 CM000362 EDX06427.1 KMY92626.1 AE013599 BT133477 AAF58869.1 AFH97167.1 AJWK01023776 GADI01003644 JAA70164.1 AY190939 CH954177 AAO01006.1 EDV58180.1 GEDC01004839 JAS32459.1 CH940648 EDW61625.1 CM000157 EDW89583.1 GEFH01004120 JAP64461.1 GDRN01077276 JAI62762.1 GACK01007520 JAA57514.1 GFPF01010772 MAA21918.1 GEDV01003152 JAP85405.1 KQ435789 KOX74464.1 GBZX01002083 JAG90657.1 BT125850 ADT78486.1 SAUD01002758 RXG69287.1 JO842277 AEO33894.1 LNIX01000033 OXA40502.1 ACPB03001854 GAHY01001844 JAA75666.1 MRZV01000333 PIK52337.1 GBBM01005652 JAC29766.1 OUUW01000001 SPP74533.1 GBSH01002066 JAG66960.1
GDQN01002196 JAT87375.1 JAT88858.1 KQ460615 KPJ13716.1 KQ459602 KPI93468.1 RSAL01000065 RVE49439.1 GAIX01003714 JAA88846.1 GALX01005852 JAB62614.1 GEZM01093128 JAV56334.1 PYGN01000202 PSN51324.1 KK852658 KDR19145.1 NEVH01017591 PNF23740.1 GAKP01004482 JAC54470.1 GDHF01032600 JAI19714.1 GFDL01006985 JAV28060.1 AXCN02001187 KQ971348 EFA04845.1 GFDG01001787 JAV17012.1 DS232051 EDS33154.1 ATLV01012285 KE524778 KFB36428.1 GAPW01004664 JAC08934.1 AAAB01008888 EAL40611.2 DQ440391 CH477693 ABF18424.1 EAT37197.1 AXCM01000042 GFDF01003846 JAV10238.1 APCN01003212 UFQS01001411 UFQT01001411 SSX10746.1 SSX30428.1 AJVK01023821 GEDC01020643 JAS16655.1 UFQS01001048 UFQT01001048 SSX08700.1 SSX28614.1 GALA01001445 JAA93407.1 KA646200 AFP60829.1 GGFM01008430 MBW29181.1 GGFM01001935 MBW22686.1 GGFJ01009556 MBW58697.1 ADMH02001608 ETN61824.1 GGFK01002546 MBW35867.1 GGFJ01009744 MBW58885.1 JRES01000505 KNC30541.1 QCYY01001600 ROT76863.1 JXJN01025954 CCAG010011323 EZ422794 ADD19070.1 CP012524 ALC42144.1 LJIJ01000340 ODM98596.1 DS235758 EEB16468.1 AJWK01022757 CH963850 EDW74723.1 GANO01002011 JAB57860.1 CH916367 EDW01453.1 CH933808 EDW08981.1 GEFM01003882 JAP71914.1 CH902619 EDV37666.1 ABJB010403120 ABJB010413655 DS761162 EEC08636.1 CM002911 KMY92627.1 CH480816 EDW47183.1 CM000362 EDX06427.1 KMY92626.1 AE013599 BT133477 AAF58869.1 AFH97167.1 AJWK01023776 GADI01003644 JAA70164.1 AY190939 CH954177 AAO01006.1 EDV58180.1 GEDC01004839 JAS32459.1 CH940648 EDW61625.1 CM000157 EDW89583.1 GEFH01004120 JAP64461.1 GDRN01077276 JAI62762.1 GACK01007520 JAA57514.1 GFPF01010772 MAA21918.1 GEDV01003152 JAP85405.1 KQ435789 KOX74464.1 GBZX01002083 JAG90657.1 BT125850 ADT78486.1 SAUD01002758 RXG69287.1 JO842277 AEO33894.1 LNIX01000033 OXA40502.1 ACPB03001854 GAHY01001844 JAA75666.1 MRZV01000333 PIK52337.1 GBBM01005652 JAC29766.1 OUUW01000001 SPP74533.1 GBSH01002066 JAG66960.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000245037
UP000027135
+ More
UP000192223 UP000091820 UP000235965 UP000075886 UP000075880 UP000007266 UP000002320 UP000075885 UP000030765 UP000075900 UP000075920 UP000075903 UP000075902 UP000076407 UP000007062 UP000008820 UP000075883 UP000075840 UP000076408 UP000075881 UP000095300 UP000092462 UP000075884 UP000095301 UP000000673 UP000069272 UP000037069 UP000283509 UP000092460 UP000092445 UP000078200 UP000092444 UP000092553 UP000094527 UP000009046 UP000092461 UP000007798 UP000001070 UP000009192 UP000007801 UP000001555 UP000001292 UP000000304 UP000000803 UP000008711 UP000008792 UP000002282 UP000192221 UP000053105 UP000288706 UP000198287 UP000015103 UP000230750 UP000268350
UP000192223 UP000091820 UP000235965 UP000075886 UP000075880 UP000007266 UP000002320 UP000075885 UP000030765 UP000075900 UP000075920 UP000075903 UP000075902 UP000076407 UP000007062 UP000008820 UP000075883 UP000075840 UP000076408 UP000075881 UP000095300 UP000092462 UP000075884 UP000095301 UP000000673 UP000069272 UP000037069 UP000283509 UP000092460 UP000092445 UP000078200 UP000092444 UP000092553 UP000094527 UP000009046 UP000092461 UP000007798 UP000001070 UP000009192 UP000007801 UP000001555 UP000001292 UP000000304 UP000000803 UP000008711 UP000008792 UP000002282 UP000192221 UP000053105 UP000288706 UP000198287 UP000015103 UP000230750 UP000268350
Pfam
PF03208 PRA1
Interpro
IPR004895
Prenylated_rab_accept_PRA1
ProteinModelPortal
A0A2A4JJY5
A0A2A4JKY1
A0A2H1VMU1
A0A1E1WPF5
A0A194RCT2
A0A194PKD3
+ More
A0A437BGA1 S4P9K8 V5GF64 A0A1Y1K4D3 A0A2P8Z497 A0A067RIM0 A0A1W4XME8 A0A1A9WP35 A0A2J7Q588 A0A034WK12 A0A0K8TZ36 A0A1Q3FKD3 A0A182QGQ3 A0A182JIB3 D2A449 A0A1L8EE76 B0WR48 A0A182P401 A0A084VEN4 A0A182RBA1 A0A023EJD0 A0A182W111 A0A182URF4 A0A182U131 A0A182WVJ3 Q5TT69 Q1HQQ3 A0A182MF79 A0A1L8DUX5 A0A182I972 A0A182YG41 A0A182K0V2 A0A336L0L8 A0A1I8PIG9 A0A1B0D3Z2 A0A1B6CTJ3 A0A336ME42 A0A182NCH2 T1E285 T1PDJ0 A0A2M3ZKX3 A0A2M3Z2E7 A0A2M4C044 W5JG84 A0A2M4A4Z3 A0A182F9K9 A0A2M4C0Q4 A0A0L0CE71 A0A3R7MAW9 A0A1B0AKK1 A0A1B0A9A8 A0A1A9UIY1 D3TMZ2 A0A0M5IXW8 A0A1D2MZY3 E0VSW2 A0A1B0CPX4 B4MRI4 U5EJZ4 B4J9H2 B4KNT6 A0A131XXD3 B3MF38 B7PPW4 A0A0J9R986 B4HMA6 B4QHU3 A1Z823 A0A1B0CQN0 A0A0K8RI51 Q8I1H0 A0A1B6E3I8 B4LJX1 B4NX22 A0A131XFI4 A0A0P4WE99 A0A1W4UZ28 L7M347 A0A224YWC8 A0A131Z3S8 A0A0N0BGF1 A0A0C9SBA7 E6PBW4 A0A444TH27 G3MK76 A0A226D4N5 R4FM62 A0A2G8KWF2 A0A023G6B3 A0A3B0J4Q5 A0A0B8RQM5
A0A437BGA1 S4P9K8 V5GF64 A0A1Y1K4D3 A0A2P8Z497 A0A067RIM0 A0A1W4XME8 A0A1A9WP35 A0A2J7Q588 A0A034WK12 A0A0K8TZ36 A0A1Q3FKD3 A0A182QGQ3 A0A182JIB3 D2A449 A0A1L8EE76 B0WR48 A0A182P401 A0A084VEN4 A0A182RBA1 A0A023EJD0 A0A182W111 A0A182URF4 A0A182U131 A0A182WVJ3 Q5TT69 Q1HQQ3 A0A182MF79 A0A1L8DUX5 A0A182I972 A0A182YG41 A0A182K0V2 A0A336L0L8 A0A1I8PIG9 A0A1B0D3Z2 A0A1B6CTJ3 A0A336ME42 A0A182NCH2 T1E285 T1PDJ0 A0A2M3ZKX3 A0A2M3Z2E7 A0A2M4C044 W5JG84 A0A2M4A4Z3 A0A182F9K9 A0A2M4C0Q4 A0A0L0CE71 A0A3R7MAW9 A0A1B0AKK1 A0A1B0A9A8 A0A1A9UIY1 D3TMZ2 A0A0M5IXW8 A0A1D2MZY3 E0VSW2 A0A1B0CPX4 B4MRI4 U5EJZ4 B4J9H2 B4KNT6 A0A131XXD3 B3MF38 B7PPW4 A0A0J9R986 B4HMA6 B4QHU3 A1Z823 A0A1B0CQN0 A0A0K8RI51 Q8I1H0 A0A1B6E3I8 B4LJX1 B4NX22 A0A131XFI4 A0A0P4WE99 A0A1W4UZ28 L7M347 A0A224YWC8 A0A131Z3S8 A0A0N0BGF1 A0A0C9SBA7 E6PBW4 A0A444TH27 G3MK76 A0A226D4N5 R4FM62 A0A2G8KWF2 A0A023G6B3 A0A3B0J4Q5 A0A0B8RQM5
Ontologies
PANTHER
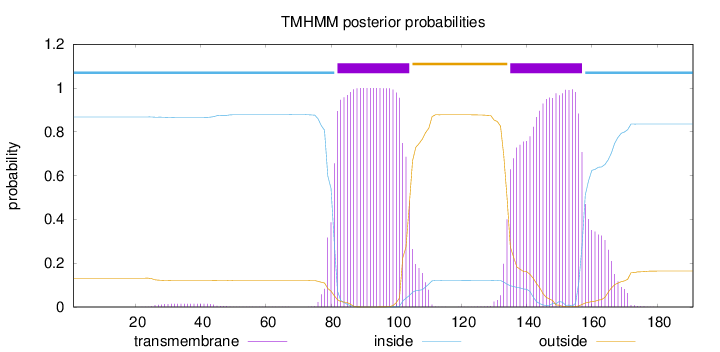
Topology
Subcellular location
Membrane
Length:
191
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
48.54989
Exp number, first 60 AAs:
0.28006
Total prob of N-in:
0.86825
inside
1 - 81
TMhelix
82 - 104
outside
105 - 134
TMhelix
135 - 157
inside
158 - 191
Population Genetic Test Statistics
Pi
31.947334
Theta
142.503697
Tajima's D
-1.718059
CLR
2165.095909
CSRT
0.0352482375881206
Interpretation
Uncertain
