Gene
KWMTBOMO09112
Annotation
PREDICTED:_uncharacterized_protein_LOC106110673_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.324
Sequence
CDS
ATGGATGAGAAATTAAAACCTATTTTAGAGGAAAATAAATACTTAAAATTAAAAGTAGAGAACTTGGAAAAGAAAATCGAAATTTTAGAGAGAGATAAGAAAACTAACAATATAATAATCCATGGATTGACAGAAGACGAGAAATCGACACCCGAGCTTATCGACAAGGTCAAAGATACAATAACAAAAGAATTAGGCATAGCAATCGAAAATTATGAATTCAACAAAATATATCGTATTGGTAAAACAAACAAAGGTAATCCATCAAAGGTAATCCGACCTATACTAATATGCTTGACTACTGGATGGAAAAAAGCGGAAATATTGAAAAATAAGAGAAAGTTAAAAGACCTGCATATCACGGAAGATTACTCTAAAGAAACCTTAGAAAAACGTAAGGCTTTACTACCACAACTCATGGAAGAACGGAGAAAAGGTAATTTCGCCTATTTAAGATATGATAAACTAATAATTAAAGAAAATAAATACGTCAAGGACTCGAGGAAGAGAGAACTCTCAATTTCACCACAAACAAACTCTCAATCCAAAAAAATTCAACCTTCAACTTCAGCCAAGAATAACAGAACGAATGCATTCGATTTGATGAGAGGTCGGTCTAACTCTTTCACAACATTGACGACAGGTCAAAAACAATAG
Protein
MDEKLKPILEENKYLKLKVENLEKKIEILERDKKTNNIIIHGLTEDEKSTPELIDKVKDTITKELGIAIENYEFNKIYRIGKTNKGNPSKVIRPILICLTTGWKKAEILKNKRKLKDLHITEDYSKETLEKRKALLPQLMEERRKGNFAYLRYDKLIIKENKYVKDSRKRELSISPQTNSQSKKIQPSTSAKNNRTNAFDLMRGRSNSFTTLTTGQKQ
Summary
Uniprot
A0A2A4JJB3
A0A2A4J1F3
A0A2A4J3C0
A0A0L7L3X8
A0A2A4IX08
A0A2A4K1M2
+ More
A0A0L7LN13 A0A0L7L7G4 A0A0L7LKR7 A0A0L7LNA7 H9JKN5 A0A2A4J436 A0A0L7KQ18 A0A2W1BGL1 A0A0L7LHJ7 A0A0L7KMI1 A0A2A4J942 A0A2H1V457 A0A2H1X3Z9 A0A2H1V475 A0A0L7LHT2 A0A0L7LNF8 A0A0L7LB65 D7F179 A0A2H1VBB6 A0A2A4JED5 A0A1E1WPR1 H9J4J3 A0A069DR53 A0A0L7LC22 A0A0L7KQ23 A0A0L7KKY9 A0A2A4KAH5 A0A1S3DLA6 A0A023FA32 A0A2H1WF27 A0A194RRA2 A0A1S4EPX8 A0A1S4EPX0 A0A0L7K3H0 A0A0L7K2J4 A0A2A4JB54 A0A0L7LJT1 A0A2A4JFH2 A0A069DRC3 A0A0L7LPK8 A0A0L7LUV7 D7F178 A0A3Q0JI53 A0A0L7K4V6 A0A1Y1K306 A0A3Q0JI62 A0A1S3DL14 A0A1S3DTW3 A0A1S4EPY6 T1I4Q9 A0A069DY11 D7F177 A0A0L7L5X6 A0A0L7L607 A0A0L7LL17 A0A3Q0IIJ5 A0A0L7KFW9 A0A1Y1LBV0 A0A1Y1MBB5 A0A0L7LNS8 A0A0L7LIU1 A0A0L7LU16 A0A0L7LLD6 A0A0L7KMI6 A0A147BCW8 A0A1Y1KNT7 A0A3Q0JIZ8 A0A1S4EA94 A0A090X8P2 A0A2H1WS99 A0A1S4ECU7
A0A0L7LN13 A0A0L7L7G4 A0A0L7LKR7 A0A0L7LNA7 H9JKN5 A0A2A4J436 A0A0L7KQ18 A0A2W1BGL1 A0A0L7LHJ7 A0A0L7KMI1 A0A2A4J942 A0A2H1V457 A0A2H1X3Z9 A0A2H1V475 A0A0L7LHT2 A0A0L7LNF8 A0A0L7LB65 D7F179 A0A2H1VBB6 A0A2A4JED5 A0A1E1WPR1 H9J4J3 A0A069DR53 A0A0L7LC22 A0A0L7KQ23 A0A0L7KKY9 A0A2A4KAH5 A0A1S3DLA6 A0A023FA32 A0A2H1WF27 A0A194RRA2 A0A1S4EPX8 A0A1S4EPX0 A0A0L7K3H0 A0A0L7K2J4 A0A2A4JB54 A0A0L7LJT1 A0A2A4JFH2 A0A069DRC3 A0A0L7LPK8 A0A0L7LUV7 D7F178 A0A3Q0JI53 A0A0L7K4V6 A0A1Y1K306 A0A3Q0JI62 A0A1S3DL14 A0A1S3DTW3 A0A1S4EPY6 T1I4Q9 A0A069DY11 D7F177 A0A0L7L5X6 A0A0L7L607 A0A0L7LL17 A0A3Q0IIJ5 A0A0L7KFW9 A0A1Y1LBV0 A0A1Y1MBB5 A0A0L7LNS8 A0A0L7LIU1 A0A0L7LU16 A0A0L7LLD6 A0A0L7KMI6 A0A147BCW8 A0A1Y1KNT7 A0A3Q0JIZ8 A0A1S4EA94 A0A090X8P2 A0A2H1WS99 A0A1S4ECU7
Pubmed
EMBL
NWSH01001196
PCG72157.1
NWSH01004020
PCG65566.1
NWSH01003397
PCG66369.1
+ More
JTDY01003098 KOB70157.1 NWSH01005231 PCG64345.1 NWSH01000260 PCG77926.1 JTDY01000539 KOB76734.1 JTDY01002536 KOB71226.1 JTDY01000743 KOB76037.1 JTDY01000511 KOB76839.1 BABH01024878 NWSH01003169 PCG66835.1 JTDY01007450 KOB65190.1 KZ150150 PZC72845.1 JTDY01001041 KOB75043.1 JTDY01008572 KOB64508.1 NWSH01002532 PCG68054.1 ODYU01000585 SOQ35630.1 ODYU01013311 SOQ60019.1 ODYU01000586 SOQ35631.1 JTDY01001071 KOB74965.1 JTDY01000484 KOB76970.1 JTDY01001949 KOB72466.1 FJ265564 ADI61832.1 ODYU01001637 SOQ38140.1 NWSH01001771 PCG70189.1 GDQN01002055 GDQN01002009 JAT88999.1 JAT89045.1 BABH01021795 GBGD01002698 JAC86191.1 JTDY01001738 KOB73038.1 JTDY01007191 KOB65392.1 JTDY01009161 KOB64038.1 NWSH01000023 PCG80652.1 GBBI01000452 JAC18260.1 ODYU01008249 SOQ51673.1 KQ459995 KPJ18566.1 JTDY01014369 KOB52037.1 JTDY01016948 KOB51874.1 NWSH01002039 PCG69337.1 JTDY01000843 KOB75692.1 NWSH01001568 PCG70835.1 GBGD01002692 JAC86197.1 JTDY01000408 KOB77334.1 JTDY01000076 KOB79001.1 FJ265563 ADI61831.1 JTDY01010650 KOB58126.1 GEZM01094174 GEZM01094173 JAV55842.1 ACPB03011704 ACPB03011705 ACPB03011706 ACPB03011707 ACPB03011708 ACPB03011709 ACPB03011710 ACPB03011711 ACPB03011712 ACPB03011713 GBGD01002635 JAC86254.1 FJ265562 ADI61830.1 JTDY01002685 KOB70933.1 JTDY01002701 KOB70892.1 JTDY01000780 JTDY01000104 JTDY01000008 KOB75906.1 KOB78842.1 KOB79479.1 JTDY01009971 KOB62006.1 GEZM01063671 JAV69026.1 GEZM01039814 JAV81166.1 JTDY01000437 KOB77173.1 JTDY01001015 KOB75121.1 JTDY01000100 KOB78869.1 JTDY01000640 KOB76373.1 JTDY01008567 KOB64512.1 GEGO01006805 JAR88599.1 GEZM01082533 JAV61335.1 GBIH01002636 JAC92074.1 ODYU01010663 SOQ55918.1
JTDY01003098 KOB70157.1 NWSH01005231 PCG64345.1 NWSH01000260 PCG77926.1 JTDY01000539 KOB76734.1 JTDY01002536 KOB71226.1 JTDY01000743 KOB76037.1 JTDY01000511 KOB76839.1 BABH01024878 NWSH01003169 PCG66835.1 JTDY01007450 KOB65190.1 KZ150150 PZC72845.1 JTDY01001041 KOB75043.1 JTDY01008572 KOB64508.1 NWSH01002532 PCG68054.1 ODYU01000585 SOQ35630.1 ODYU01013311 SOQ60019.1 ODYU01000586 SOQ35631.1 JTDY01001071 KOB74965.1 JTDY01000484 KOB76970.1 JTDY01001949 KOB72466.1 FJ265564 ADI61832.1 ODYU01001637 SOQ38140.1 NWSH01001771 PCG70189.1 GDQN01002055 GDQN01002009 JAT88999.1 JAT89045.1 BABH01021795 GBGD01002698 JAC86191.1 JTDY01001738 KOB73038.1 JTDY01007191 KOB65392.1 JTDY01009161 KOB64038.1 NWSH01000023 PCG80652.1 GBBI01000452 JAC18260.1 ODYU01008249 SOQ51673.1 KQ459995 KPJ18566.1 JTDY01014369 KOB52037.1 JTDY01016948 KOB51874.1 NWSH01002039 PCG69337.1 JTDY01000843 KOB75692.1 NWSH01001568 PCG70835.1 GBGD01002692 JAC86197.1 JTDY01000408 KOB77334.1 JTDY01000076 KOB79001.1 FJ265563 ADI61831.1 JTDY01010650 KOB58126.1 GEZM01094174 GEZM01094173 JAV55842.1 ACPB03011704 ACPB03011705 ACPB03011706 ACPB03011707 ACPB03011708 ACPB03011709 ACPB03011710 ACPB03011711 ACPB03011712 ACPB03011713 GBGD01002635 JAC86254.1 FJ265562 ADI61830.1 JTDY01002685 KOB70933.1 JTDY01002701 KOB70892.1 JTDY01000780 JTDY01000104 JTDY01000008 KOB75906.1 KOB78842.1 KOB79479.1 JTDY01009971 KOB62006.1 GEZM01063671 JAV69026.1 GEZM01039814 JAV81166.1 JTDY01000437 KOB77173.1 JTDY01001015 KOB75121.1 JTDY01000100 KOB78869.1 JTDY01000640 KOB76373.1 JTDY01008567 KOB64512.1 GEGO01006805 JAR88599.1 GEZM01082533 JAV61335.1 GBIH01002636 JAC92074.1 ODYU01010663 SOQ55918.1
Proteomes
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR027124 Swc5/CFDP2
IPR000477 RT_dom
IPR004244 Transposase_22
IPR020845 AMP-binding_CS
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR036691 Endo/exonu/phosph_ase_sf
IPR027124 Swc5/CFDP2
IPR000477 RT_dom
IPR004244 Transposase_22
IPR020845 AMP-binding_CS
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR036322 WD40_repeat_dom_sf
Gene 3D
ProteinModelPortal
A0A2A4JJB3
A0A2A4J1F3
A0A2A4J3C0
A0A0L7L3X8
A0A2A4IX08
A0A2A4K1M2
+ More
A0A0L7LN13 A0A0L7L7G4 A0A0L7LKR7 A0A0L7LNA7 H9JKN5 A0A2A4J436 A0A0L7KQ18 A0A2W1BGL1 A0A0L7LHJ7 A0A0L7KMI1 A0A2A4J942 A0A2H1V457 A0A2H1X3Z9 A0A2H1V475 A0A0L7LHT2 A0A0L7LNF8 A0A0L7LB65 D7F179 A0A2H1VBB6 A0A2A4JED5 A0A1E1WPR1 H9J4J3 A0A069DR53 A0A0L7LC22 A0A0L7KQ23 A0A0L7KKY9 A0A2A4KAH5 A0A1S3DLA6 A0A023FA32 A0A2H1WF27 A0A194RRA2 A0A1S4EPX8 A0A1S4EPX0 A0A0L7K3H0 A0A0L7K2J4 A0A2A4JB54 A0A0L7LJT1 A0A2A4JFH2 A0A069DRC3 A0A0L7LPK8 A0A0L7LUV7 D7F178 A0A3Q0JI53 A0A0L7K4V6 A0A1Y1K306 A0A3Q0JI62 A0A1S3DL14 A0A1S3DTW3 A0A1S4EPY6 T1I4Q9 A0A069DY11 D7F177 A0A0L7L5X6 A0A0L7L607 A0A0L7LL17 A0A3Q0IIJ5 A0A0L7KFW9 A0A1Y1LBV0 A0A1Y1MBB5 A0A0L7LNS8 A0A0L7LIU1 A0A0L7LU16 A0A0L7LLD6 A0A0L7KMI6 A0A147BCW8 A0A1Y1KNT7 A0A3Q0JIZ8 A0A1S4EA94 A0A090X8P2 A0A2H1WS99 A0A1S4ECU7
A0A0L7LN13 A0A0L7L7G4 A0A0L7LKR7 A0A0L7LNA7 H9JKN5 A0A2A4J436 A0A0L7KQ18 A0A2W1BGL1 A0A0L7LHJ7 A0A0L7KMI1 A0A2A4J942 A0A2H1V457 A0A2H1X3Z9 A0A2H1V475 A0A0L7LHT2 A0A0L7LNF8 A0A0L7LB65 D7F179 A0A2H1VBB6 A0A2A4JED5 A0A1E1WPR1 H9J4J3 A0A069DR53 A0A0L7LC22 A0A0L7KQ23 A0A0L7KKY9 A0A2A4KAH5 A0A1S3DLA6 A0A023FA32 A0A2H1WF27 A0A194RRA2 A0A1S4EPX8 A0A1S4EPX0 A0A0L7K3H0 A0A0L7K2J4 A0A2A4JB54 A0A0L7LJT1 A0A2A4JFH2 A0A069DRC3 A0A0L7LPK8 A0A0L7LUV7 D7F178 A0A3Q0JI53 A0A0L7K4V6 A0A1Y1K306 A0A3Q0JI62 A0A1S3DL14 A0A1S3DTW3 A0A1S4EPY6 T1I4Q9 A0A069DY11 D7F177 A0A0L7L5X6 A0A0L7L607 A0A0L7LL17 A0A3Q0IIJ5 A0A0L7KFW9 A0A1Y1LBV0 A0A1Y1MBB5 A0A0L7LNS8 A0A0L7LIU1 A0A0L7LU16 A0A0L7LLD6 A0A0L7KMI6 A0A147BCW8 A0A1Y1KNT7 A0A3Q0JIZ8 A0A1S4EA94 A0A090X8P2 A0A2H1WS99 A0A1S4ECU7
Ontologies
Topology
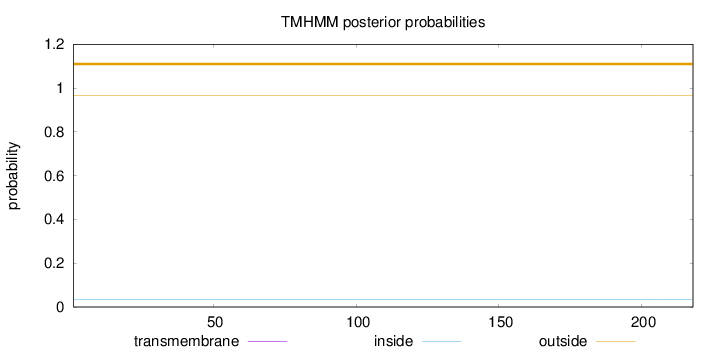
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00123
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03521
outside
1 - 218
Population Genetic Test Statistics
Pi
227.011546
Theta
129.386782
Tajima's D
2.162949
CLR
2.474032
CSRT
0.906954652267387
Interpretation
Uncertain
