Gene
KWMTBOMO09106 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007571
Annotation
PREDICTED:_eukaryotic_translation_initiation_factor_2A_[Amyelois_transitella]
Full name
Eukaryotic translation initiation factor 2A
Alternative Name
65 kDa eukaryotic translation initiation factor 2A
Location in the cell
Mitochondrial Reliability : 1.99
Sequence
CDS
ATGACTAACCCGCATGTCGCAGGTCTAACCAATAAAAGTATATTCTTGCTTGAGTTTAAAGAGCCATATTGTGAGAAAACCTTAAGAGATGAATCAACAACAAATGTACGAGGCATTATATGTAGCCCTAAAGGGTCTTACTTGGCCTATAGAACGGATAGAAAATCTGAGATAGTCAGATGCTCAGACTGGTCTACGACAGCATCAATTGATGGAAACGTTAAGGAGATGTTTTTCTCTCCATTGGACAACTTTTTTATGGTTTGGGAAATGTATGCCATCACTAAAGATAATCCTGAGGGTAAACCAAACCTCAGAGTATTCAAAACAGATAATGGTAAATTAGTTGGATCATTTGTACAGAAAAATCAAATAGGATGGGAGCCAAAATGGTCTACGGATGAGAAGTTATTTGCCATTCAATTAAATAATAAAATCCAGATATATGAAGATGGGAACTTAGAACGCTATGTCCATCAATACAGTGCAGAAAAATTAAAGGCGTTCAGTCTCTCGCCGAGTCCTTCTCCGAATTATTACATTGCTGTATTTACATTAGGTAAACAAGGGCAGCCATCATTTTGGCGTGTATTTAAATACCCTCAATTTGATGTTAGTCAAGCTGTTGTCACCCGGTCATCCTTCCAAGCTGATAAGGCTGTGTTCCACTGGAACAGACGCGGTACAAATGTGTTTGCTATGACACAGACAGATGTTGACAAAACGGGCGGTTCGTACTACGGAAAACAAACGCTATCGCATGGCGACGTCAAAGGGAACTCTGGAATCGTATCGTTCAGTAAGGAAGGTCCCATCCACGCGATTAGTTGGAACCCGGGCAACTGGAACGAGTGGATAGCGGTGTACGGTCACGCCCCGGCCAAAGCGACGGTCTTCAATGCGAAATGCGAACCTCTTTTCGAGTTTGGCACCGGAGCTTGGAACTCGATTTACTTCTCCCCCGAAGGAACTCATGTCCTTTTGGGAGGCTTTGGGAACATAGCGACCGGCCACATCGAGGTGTGGGACATGCGCGGGTACAAAAAGGCGGGCACTTGTTCCGCTCCAGATACAACTTTCCTGCAATGGGACCCGAGGGGTGAAACCTTCCTGACTGCTACTACTTACCCTAGGCTCACTGTTAGCAATGGATATAAAATATGGCATTACACGGGCGTATTGATGCACAAGCGTTCGTGTAGCGAGAAGGAGAATCTTCTGGCAATATGCTGGCAGCCTGGATCATATCCCAAAACGGATCTATCGAAGAGCGCTCCTAAGGCTATCGAGGACTCAACGCCGAAGCCAATGGGCGCCTATCGCCCCCCAGGAGCTCGGAGCAGACCTTCAAACTTTACGCTACACGAACACGAGAAACCTCATAAACCTGGAGAGAGTCCTGCTGTTGGACCGTCAAAGCAAGCGATCAAGCAGAAAGAGAAACGTGAAGCTCGAAAAGCTCGTAAAGCTGAAGAAAAGGGGAAAGGCGACGTGTCCCCGGAAGATGGGTCCTCGCCGGTCCCCCCCACCACTACACGGCAACCCTTCGTCTCGACGGGCGATCCCGAAAAGGATAAGAAAATCAAGAACATCAACAAGAAATTAAACGACATAGACAAATTGAAACAGCAAAAAGCCGCCGGGAAGCCGCTAGAGTTGAACCAACTCGCCAAGATCGACAACGAACAAGCATTACTGCAAGAATTGTCTCAGCTGAAGTTATGA
Protein
MTNPHVAGLTNKSIFLLEFKEPYCEKTLRDESTTNVRGIICSPKGSYLAYRTDRKSEIVRCSDWSTTASIDGNVKEMFFSPLDNFFMVWEMYAITKDNPEGKPNLRVFKTDNGKLVGSFVQKNQIGWEPKWSTDEKLFAIQLNNKIQIYEDGNLERYVHQYSAEKLKAFSLSPSPSPNYYIAVFTLGKQGQPSFWRVFKYPQFDVSQAVVTRSSFQADKAVFHWNRRGTNVFAMTQTDVDKTGGSYYGKQTLSHGDVKGNSGIVSFSKEGPIHAISWNPGNWNEWIAVYGHAPAKATVFNAKCEPLFEFGTGAWNSIYFSPEGTHVLLGGFGNIATGHIEVWDMRGYKKAGTCSAPDTTFLQWDPRGETFLTATTYPRLTVSNGYKIWHYTGVLMHKRSCSEKENLLAICWQPGSYPKTDLSKSAPKAIEDSTPKPMGAYRPPGARSRPSNFTLHEHEKPHKPGESPAVGPSKQAIKQKEKREARKARKAEEKGKGDVSPEDGSSPVPPTTTRQPFVSTGDPEKDKKIKNINKKLNDIDKLKQQKAAGKPLELNQLAKIDNEQALLQELSQLKL
Summary
Description
Functions in the early steps of protein synthesis of a small number of specific mRNAs. Acts by directing the binding of methionyl-tRNAi to 40S ribosomal subunits. In contrast to the eIF-2 complex, it binds methionyl-tRNAi to 40S subunits in a codon-dependent manner, whereas the eIF-2 complex binds methionyl-tRNAi to 40S subunits in a GTP-dependent manner.
Similarity
Belongs to the WD repeat EIF2A family.
Keywords
Coiled coil
Complete proteome
Initiation factor
Protein biosynthesis
Reference proteome
Repeat
Translation regulation
WD repeat
Acetylation
Alternative splicing
Direct protein sequencing
Phosphoprotein
Polymorphism
Feature
chain Eukaryotic translation initiation factor 2A
splice variant In isoform 2.
sequence variant In dbSNP:rs1132979.
splice variant In isoform 2.
sequence variant In dbSNP:rs1132979.
Uniprot
S4PJP5
A0A2A4K3G1
A0A437BG91
A0A194R8X6
A0A2H1WNI0
D6WL38
+ More
A0A232F8Z3 J3JVJ2 U4UPA3 A0A1W4X5H6 N6TMA7 A0A3L8DR28 A0A026WBK5 A0A158NJ56 A0A067RBG5 A0A088A5Z7 A0A0C9R134 A0A3B3SYS8 A0A1B6FTV9 A0A146NU77 A0A0K8SU85 A0A0A9WRR4 A0A3Q2QGX4 A0A3Q2C732 A0A3P8TLP2 A0A154P5M1 A0A1B6DER9 A0A3Q2T1S1 A0A3Q1AQ97 E9J1W3 A0A310SE97 A0A2Y9K6F0 A0A151JRM4 A0A1A8UZ93 A0A2K6QR05 A0A3P4RXD2 I3KL04 A0A2Y9JVU3 G3ST16 A0A0P4ZEA3 M3XU13 A0A0P5INM1 A0A0P5HTJ9 Q5ZKC1 A0A0P5H4Y6 A0A1A7ZYI5 A0A1A8DYX9 A0A1L1RYW0 A0A0P5PVP3 A0A0P5J0V7 A0A0P5SYA3 G3TZV7 A0A2K6LZJ4 A0A0P5SC65 A0A0P5B3Z1 A0A094L1U7 A0A093GHU5 A0A1Y1LMN8 A0A340XA42 A0A2K6QR10 A0A0A0AF29 A0A3Q2P5M9 A0A401SIW9 A0A1A8J2I6 A0A2I0TZU6 A0A091H4G0 A0A2K6LZK5 A0A3B3VQP9 A0A2I2V1V6 M3WEX5 A0A452FUT9 A0A3P8PBZ3 A0A3P9AXB8 A0A452FUY7 A0A452FUZ7 A0A1A8QL46 A0A1A8NJA5 A0A0N8AGY0 W5P328 A0A3B1IFN3 A0A1B0DCV7 A0A091WMI5 A0A3Q7R7E0 A0A250Y325 S7PDP4 E9FV01 I3M265 Q9BY44 A0A2K6SVD1 A0A1A8ER44 A0A091EQB9 A0A091L451 A0A0Q3TIZ1 F1PGT8 A0A3Q7U1J4 G1NEN3 A0A2K5F811 A0A2K5F7W4
A0A232F8Z3 J3JVJ2 U4UPA3 A0A1W4X5H6 N6TMA7 A0A3L8DR28 A0A026WBK5 A0A158NJ56 A0A067RBG5 A0A088A5Z7 A0A0C9R134 A0A3B3SYS8 A0A1B6FTV9 A0A146NU77 A0A0K8SU85 A0A0A9WRR4 A0A3Q2QGX4 A0A3Q2C732 A0A3P8TLP2 A0A154P5M1 A0A1B6DER9 A0A3Q2T1S1 A0A3Q1AQ97 E9J1W3 A0A310SE97 A0A2Y9K6F0 A0A151JRM4 A0A1A8UZ93 A0A2K6QR05 A0A3P4RXD2 I3KL04 A0A2Y9JVU3 G3ST16 A0A0P4ZEA3 M3XU13 A0A0P5INM1 A0A0P5HTJ9 Q5ZKC1 A0A0P5H4Y6 A0A1A7ZYI5 A0A1A8DYX9 A0A1L1RYW0 A0A0P5PVP3 A0A0P5J0V7 A0A0P5SYA3 G3TZV7 A0A2K6LZJ4 A0A0P5SC65 A0A0P5B3Z1 A0A094L1U7 A0A093GHU5 A0A1Y1LMN8 A0A340XA42 A0A2K6QR10 A0A0A0AF29 A0A3Q2P5M9 A0A401SIW9 A0A1A8J2I6 A0A2I0TZU6 A0A091H4G0 A0A2K6LZK5 A0A3B3VQP9 A0A2I2V1V6 M3WEX5 A0A452FUT9 A0A3P8PBZ3 A0A3P9AXB8 A0A452FUY7 A0A452FUZ7 A0A1A8QL46 A0A1A8NJA5 A0A0N8AGY0 W5P328 A0A3B1IFN3 A0A1B0DCV7 A0A091WMI5 A0A3Q7R7E0 A0A250Y325 S7PDP4 E9FV01 I3M265 Q9BY44 A0A2K6SVD1 A0A1A8ER44 A0A091EQB9 A0A091L451 A0A0Q3TIZ1 F1PGT8 A0A3Q7U1J4 G1NEN3 A0A2K5F811 A0A2K5F7W4
Pubmed
23622113
26354079
18362917
19820115
28648823
22516182
+ More
23537049 30249741 24508170 21347285 24845553 29240929 25401762 26823975 21282665 25362486 25186727 15642098 15592404 28004739 30297745 17975172 20809919 25329095 28087693 21292972 12133843 14702039 16641997 15489334 17081983 18669648 19413330 19690332 20068231 21269460 21406692 22814378 23186163 24275569 16341006 20838655
23537049 30249741 24508170 21347285 24845553 29240929 25401762 26823975 21282665 25362486 25186727 15642098 15592404 28004739 30297745 17975172 20809919 25329095 28087693 21292972 12133843 14702039 16641997 15489334 17081983 18669648 19413330 19690332 20068231 21269460 21406692 22814378 23186163 24275569 16341006 20838655
EMBL
GAIX01004865
JAA87695.1
NWSH01000191
PCG78589.1
RSAL01000065
RVE49429.1
+ More
KQ460615 KPJ13705.1 ODYU01009822 SOQ54502.1 KQ971343 EFA03515.1 NNAY01000700 OXU26913.1 BT127260 AEE62222.1 KB632303 ERL91840.1 APGK01018933 KB740085 ENN81579.1 QOIP01000005 RLU22894.1 KK107293 EZA53440.1 ADTU01017494 KK852751 KDR17144.1 GBYB01006612 JAG76379.1 GECZ01016112 JAS53657.1 GCES01151596 JAQ34726.1 GBRD01008958 JAG56863.1 GBHO01036044 GDHC01018224 JAG07560.1 JAQ00405.1 KQ434809 KZC06488.1 GEDC01029044 GEDC01013100 JAS08254.1 JAS24198.1 GL767674 EFZ13269.1 KQ760706 OAD59323.1 KQ978579 KYN30006.1 HAEJ01012948 SBS53405.1 CYRY02044596 VCX39604.1 AERX01047804 AERX01047805 GDIP01215430 JAJ07972.1 AEYP01039192 AEYP01039193 AEYP01039194 AEYP01039195 AEYP01039196 AEYP01039197 GDIQ01212695 JAK39030.1 GDIQ01228943 JAK22782.1 AJ720163 GDIQ01238988 JAK12737.1 HADY01008968 SBP47453.1 HADZ01010323 HAEA01009519 SBQ37999.1 AADN05000526 GDIQ01125080 JAL26646.1 GDIQ01231193 JAK20532.1 GDIQ01098741 JAL52985.1 GDIP01141653 JAL62061.1 GDIP01189978 JAJ33424.1 KL360001 KFZ64805.1 KL216281 KFV68793.1 GEZM01051462 GEZM01051461 JAV74912.1 KL871660 KGL92766.1 BEZZ01000298 GCC30354.1 HAED01017382 HAEE01006454 SBR03827.1 KZ506511 PKU39356.1 KL448192 KFO81182.1 AANG04001816 LWLT01000001 HAEH01004536 HAEI01005653 SBR94261.1 HAEF01001712 HAEG01003269 SBR68897.1 GDIP01148497 JAJ74905.1 AMGL01010877 AJVK01014242 KK736027 KFR16857.1 GFFV01001908 JAV38037.1 KE161952 EPQ06037.1 GL732525 EFX88811.1 AGTP01103865 AGTP01103866 AF497978 AK027356 AK298586 AK293993 AF212241 AF109358 AF172818 AC107426 CH471052 BC011885 HAEB01002810 SBQ49337.1 KK718888 KFO59221.1 KL300671 KFP51209.1 LMAW01002523 KQK80542.1 AAEX03013679 AAEX03013680
KQ460615 KPJ13705.1 ODYU01009822 SOQ54502.1 KQ971343 EFA03515.1 NNAY01000700 OXU26913.1 BT127260 AEE62222.1 KB632303 ERL91840.1 APGK01018933 KB740085 ENN81579.1 QOIP01000005 RLU22894.1 KK107293 EZA53440.1 ADTU01017494 KK852751 KDR17144.1 GBYB01006612 JAG76379.1 GECZ01016112 JAS53657.1 GCES01151596 JAQ34726.1 GBRD01008958 JAG56863.1 GBHO01036044 GDHC01018224 JAG07560.1 JAQ00405.1 KQ434809 KZC06488.1 GEDC01029044 GEDC01013100 JAS08254.1 JAS24198.1 GL767674 EFZ13269.1 KQ760706 OAD59323.1 KQ978579 KYN30006.1 HAEJ01012948 SBS53405.1 CYRY02044596 VCX39604.1 AERX01047804 AERX01047805 GDIP01215430 JAJ07972.1 AEYP01039192 AEYP01039193 AEYP01039194 AEYP01039195 AEYP01039196 AEYP01039197 GDIQ01212695 JAK39030.1 GDIQ01228943 JAK22782.1 AJ720163 GDIQ01238988 JAK12737.1 HADY01008968 SBP47453.1 HADZ01010323 HAEA01009519 SBQ37999.1 AADN05000526 GDIQ01125080 JAL26646.1 GDIQ01231193 JAK20532.1 GDIQ01098741 JAL52985.1 GDIP01141653 JAL62061.1 GDIP01189978 JAJ33424.1 KL360001 KFZ64805.1 KL216281 KFV68793.1 GEZM01051462 GEZM01051461 JAV74912.1 KL871660 KGL92766.1 BEZZ01000298 GCC30354.1 HAED01017382 HAEE01006454 SBR03827.1 KZ506511 PKU39356.1 KL448192 KFO81182.1 AANG04001816 LWLT01000001 HAEH01004536 HAEI01005653 SBR94261.1 HAEF01001712 HAEG01003269 SBR68897.1 GDIP01148497 JAJ74905.1 AMGL01010877 AJVK01014242 KK736027 KFR16857.1 GFFV01001908 JAV38037.1 KE161952 EPQ06037.1 GL732525 EFX88811.1 AGTP01103865 AGTP01103866 AF497978 AK027356 AK298586 AK293993 AF212241 AF109358 AF172818 AC107426 CH471052 BC011885 HAEB01002810 SBQ49337.1 KK718888 KFO59221.1 KL300671 KFP51209.1 LMAW01002523 KQK80542.1 AAEX03013679 AAEX03013680
Proteomes
UP000218220
UP000283053
UP000053240
UP000007266
UP000215335
UP000030742
+ More
UP000192223 UP000019118 UP000279307 UP000053097 UP000005205 UP000027135 UP000005203 UP000261540 UP000265000 UP000265020 UP000265080 UP000076502 UP000257160 UP000248482 UP000078492 UP000233200 UP000005207 UP000007646 UP000000715 UP000000539 UP000233180 UP000053875 UP000265300 UP000053858 UP000287033 UP000053760 UP000261500 UP000011712 UP000291000 UP000265100 UP000265160 UP000002356 UP000018467 UP000092462 UP000053605 UP000286641 UP000000305 UP000005215 UP000005640 UP000233220 UP000052976 UP000051836 UP000002254 UP000286640 UP000001645 UP000233020
UP000192223 UP000019118 UP000279307 UP000053097 UP000005205 UP000027135 UP000005203 UP000261540 UP000265000 UP000265020 UP000265080 UP000076502 UP000257160 UP000248482 UP000078492 UP000233200 UP000005207 UP000007646 UP000000715 UP000000539 UP000233180 UP000053875 UP000265300 UP000053858 UP000287033 UP000053760 UP000261500 UP000011712 UP000291000 UP000265100 UP000265160 UP000002356 UP000018467 UP000092462 UP000053605 UP000286641 UP000000305 UP000005215 UP000005640 UP000233220 UP000052976 UP000051836 UP000002254 UP000286640 UP000001645 UP000233020
Pfam
PF08662 eIF2A
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
S4PJP5
A0A2A4K3G1
A0A437BG91
A0A194R8X6
A0A2H1WNI0
D6WL38
+ More
A0A232F8Z3 J3JVJ2 U4UPA3 A0A1W4X5H6 N6TMA7 A0A3L8DR28 A0A026WBK5 A0A158NJ56 A0A067RBG5 A0A088A5Z7 A0A0C9R134 A0A3B3SYS8 A0A1B6FTV9 A0A146NU77 A0A0K8SU85 A0A0A9WRR4 A0A3Q2QGX4 A0A3Q2C732 A0A3P8TLP2 A0A154P5M1 A0A1B6DER9 A0A3Q2T1S1 A0A3Q1AQ97 E9J1W3 A0A310SE97 A0A2Y9K6F0 A0A151JRM4 A0A1A8UZ93 A0A2K6QR05 A0A3P4RXD2 I3KL04 A0A2Y9JVU3 G3ST16 A0A0P4ZEA3 M3XU13 A0A0P5INM1 A0A0P5HTJ9 Q5ZKC1 A0A0P5H4Y6 A0A1A7ZYI5 A0A1A8DYX9 A0A1L1RYW0 A0A0P5PVP3 A0A0P5J0V7 A0A0P5SYA3 G3TZV7 A0A2K6LZJ4 A0A0P5SC65 A0A0P5B3Z1 A0A094L1U7 A0A093GHU5 A0A1Y1LMN8 A0A340XA42 A0A2K6QR10 A0A0A0AF29 A0A3Q2P5M9 A0A401SIW9 A0A1A8J2I6 A0A2I0TZU6 A0A091H4G0 A0A2K6LZK5 A0A3B3VQP9 A0A2I2V1V6 M3WEX5 A0A452FUT9 A0A3P8PBZ3 A0A3P9AXB8 A0A452FUY7 A0A452FUZ7 A0A1A8QL46 A0A1A8NJA5 A0A0N8AGY0 W5P328 A0A3B1IFN3 A0A1B0DCV7 A0A091WMI5 A0A3Q7R7E0 A0A250Y325 S7PDP4 E9FV01 I3M265 Q9BY44 A0A2K6SVD1 A0A1A8ER44 A0A091EQB9 A0A091L451 A0A0Q3TIZ1 F1PGT8 A0A3Q7U1J4 G1NEN3 A0A2K5F811 A0A2K5F7W4
A0A232F8Z3 J3JVJ2 U4UPA3 A0A1W4X5H6 N6TMA7 A0A3L8DR28 A0A026WBK5 A0A158NJ56 A0A067RBG5 A0A088A5Z7 A0A0C9R134 A0A3B3SYS8 A0A1B6FTV9 A0A146NU77 A0A0K8SU85 A0A0A9WRR4 A0A3Q2QGX4 A0A3Q2C732 A0A3P8TLP2 A0A154P5M1 A0A1B6DER9 A0A3Q2T1S1 A0A3Q1AQ97 E9J1W3 A0A310SE97 A0A2Y9K6F0 A0A151JRM4 A0A1A8UZ93 A0A2K6QR05 A0A3P4RXD2 I3KL04 A0A2Y9JVU3 G3ST16 A0A0P4ZEA3 M3XU13 A0A0P5INM1 A0A0P5HTJ9 Q5ZKC1 A0A0P5H4Y6 A0A1A7ZYI5 A0A1A8DYX9 A0A1L1RYW0 A0A0P5PVP3 A0A0P5J0V7 A0A0P5SYA3 G3TZV7 A0A2K6LZJ4 A0A0P5SC65 A0A0P5B3Z1 A0A094L1U7 A0A093GHU5 A0A1Y1LMN8 A0A340XA42 A0A2K6QR10 A0A0A0AF29 A0A3Q2P5M9 A0A401SIW9 A0A1A8J2I6 A0A2I0TZU6 A0A091H4G0 A0A2K6LZK5 A0A3B3VQP9 A0A2I2V1V6 M3WEX5 A0A452FUT9 A0A3P8PBZ3 A0A3P9AXB8 A0A452FUY7 A0A452FUZ7 A0A1A8QL46 A0A1A8NJA5 A0A0N8AGY0 W5P328 A0A3B1IFN3 A0A1B0DCV7 A0A091WMI5 A0A3Q7R7E0 A0A250Y325 S7PDP4 E9FV01 I3M265 Q9BY44 A0A2K6SVD1 A0A1A8ER44 A0A091EQB9 A0A091L451 A0A0Q3TIZ1 F1PGT8 A0A3Q7U1J4 G1NEN3 A0A2K5F811 A0A2K5F7W4
PDB
3WJ9
E-value=1.4233e-53,
Score=532
Ontologies
GO
PANTHER
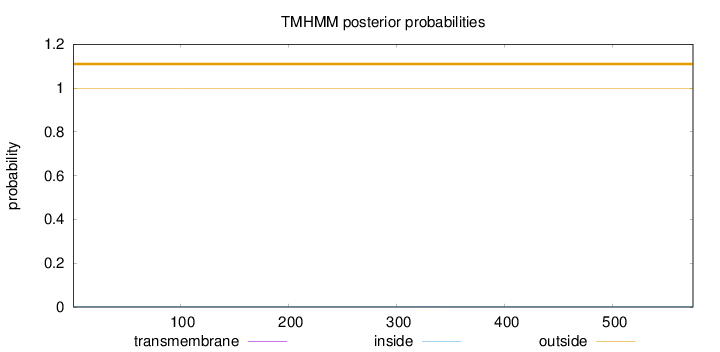
Topology
Length:
574
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00941999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00073
outside
1 - 574
Population Genetic Test Statistics
Pi
200.934158
Theta
146.483776
Tajima's D
0.420778
CLR
1.240975
CSRT
0.494475276236188
Interpretation
Uncertain
