Gene
KWMTBOMO09102
Pre Gene Modal
BGIBMGA007525
Annotation
PREDICTED:_uncharacterized_protein_LOC101742533_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.003 Extracellular Reliability : 1.034 Nuclear Reliability : 1.658
Sequence
CDS
ATGTTTCTGACGACCATTGTAATACTTGTCTCAACGCTGAGAGTTCAAGGGTTCTGCGGCGATAGTATTCGCTGGTCGCACCATTTCAAGATGGACGATGTCTTCGGTATGTGGTACGGAGCGGGGTACGCGCAGCACACACCTGACATGACTAATAAACCTAACGAAATTGGTTGCGTCACTTTGTACATTACTGACGTGACCACGGAGCCCCGCGAAGACTGGTTGGATTGGTCCATTATCCGGCCTAACTATTCCGATTACAATTGGCGGTCGTACAAGAGTAATCCGTGGTCAGATAATTCAATGTCCGGATCTTGGGTTGATGTTAAAATCAAGAGAAGCGTTCGGAGAGACATGTACAGTGAGAGACGTCTGCGAATTATTTGGGATGAAGATGGTCAAAGTATGGAGCAGACGTATGAATATTCGACTGACGAACCTGGTTTATGGGTTGCTGAGGATAGAAGACAGCTTGAAAGAGAAATGCGAGCTAGAGGAATGGATATTTGGGATCCAAACGTGCTTCCGAGACATCCGCAAGTAATAAGAATCCTAAAAGCATCTCAGCATTTATTGATCATCAATCATTGTTCGGAAATAGGTGACGGTGGTATTTTCACGCTTATCTTGAGGAGATCACCATCAAAAATGAATAGGTATAATGTTGGCGCCGATGATAGTGCCGATGGCGTTCATTAA
Protein
MFLTTIVILVSTLRVQGFCGDSIRWSHHFKMDDVFGMWYGAGYAQHTPDMTNKPNEIGCVTLYITDVTTEPREDWLDWSIIRPNYSDYNWRSYKSNPWSDNSMSGSWVDVKIKRSVRRDMYSERRLRIIWDEDGQSMEQTYEYSTDEPGLWVAEDRRQLEREMRARGMDIWDPNVLPRHPQVIRILKASQHLLIINHCSEIGDGGIFTLILRRSPSKMNRYNVGADDSADGVH
Summary
Uniprot
H9JDC8
A0A0L7L0K2
A0A2H1X164
A0A437BG64
A0A2A4K3U8
A0A194PQP4
+ More
A0A194RCR7 A0A212F032 A0A182PBA8 A0A182NSQ5 A0A182I8Q1 A0A182QT35 A0A182U7L9 A0A182LAL0 Q7QFU6 A0A182WV25 A0A1I8QBA5 A0A0M4E9Z5 A0A182URT5 A0A0M3QW56 B4PFE6 B3NFP6 B4N702 A0A182XWN7 A0A1W4V043 A0A182VY76 A0A182RDB8 A0A0L0C4T5 B4HKK9 A0A182JDM6 B4QMQ1 Q8IQB7 Q8IGE3 T1P8D8 A0A1I8MMX4 A0A023EM12 A0A0K8WGU4 A0A2M3ZEH9 B3M8W3 A0A084WNT3 A0A1J1IVG7 A0A1A9W7L7 A0A0A1XM50 H9JDC9 A0A034W876 A0A437BG67 A0A182GB94 B4LB45 D7EJD9 A0A3B0JWC0 Q29F54 B4HB05 A0A1Y1MWF7 B4J0X4 A0A194R7S7 A0A2A4K2V2 A0A3B0JTD7 B4L9H6 A0A2M4BUH0 A0A2M4BUJ8 A0A2M4BUQ9 A0A1Q3FP71 B4LAN8 A0A1A9VEA4 A0A1L8DP78 A0A182F7U6 A0A1L8DPE1 A0A336JY31
A0A194RCR7 A0A212F032 A0A182PBA8 A0A182NSQ5 A0A182I8Q1 A0A182QT35 A0A182U7L9 A0A182LAL0 Q7QFU6 A0A182WV25 A0A1I8QBA5 A0A0M4E9Z5 A0A182URT5 A0A0M3QW56 B4PFE6 B3NFP6 B4N702 A0A182XWN7 A0A1W4V043 A0A182VY76 A0A182RDB8 A0A0L0C4T5 B4HKK9 A0A182JDM6 B4QMQ1 Q8IQB7 Q8IGE3 T1P8D8 A0A1I8MMX4 A0A023EM12 A0A0K8WGU4 A0A2M3ZEH9 B3M8W3 A0A084WNT3 A0A1J1IVG7 A0A1A9W7L7 A0A0A1XM50 H9JDC9 A0A034W876 A0A437BG67 A0A182GB94 B4LB45 D7EJD9 A0A3B0JWC0 Q29F54 B4HB05 A0A1Y1MWF7 B4J0X4 A0A194R7S7 A0A2A4K2V2 A0A3B0JTD7 B4L9H6 A0A2M4BUH0 A0A2M4BUJ8 A0A2M4BUQ9 A0A1Q3FP71 B4LAN8 A0A1A9VEA4 A0A1L8DP78 A0A182F7U6 A0A1L8DPE1 A0A336JY31
Pubmed
19121390
26227816
26354079
22118469
20966253
12364791
+ More
14747013 17210077 17994087 17550304 25244985 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 24438588 25830018 25348373 26483478 18362917 19820115 15632085 28004739
14747013 17210077 17994087 17550304 25244985 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24945155 24438588 25830018 25348373 26483478 18362917 19820115 15632085 28004739
EMBL
BABH01030112
JTDY01003909
KOB68841.1
ODYU01012656
SOQ59085.1
RSAL01000065
+ More
RVE49426.1 NWSH01000191 PCG78578.1 KQ459602 KPI93450.1 KQ460615 KPJ13701.1 AGBW02011160 OWR47115.1 APCN01003099 AXCN02000230 AAAB01008844 EAA05949.4 CP012525 ALC44004.1 ALC43563.1 CM000159 EDW93072.1 CH954178 EDV50588.2 CH964168 EDW80141.2 JRES01000919 KNC27262.1 CH480815 EDW40812.1 CM000363 CM002912 EDX09805.1 KMY98558.1 AE014296 BT128694 AAN11985.1 AEE69542.1 BT001823 AAN71578.1 KA644894 AFP59523.1 GAPW01003265 JAC10333.1 GDHF01002050 JAI50264.1 GGFM01006158 MBW26909.1 CH902618 EDV41114.1 ATLV01024686 KE525357 KFB51877.1 CVRI01000062 CRL04205.1 GBXI01001903 JAD12389.1 BABH01030111 GAKP01008969 JAC49983.1 RVE49427.1 JXUM01052337 KQ561729 KXJ77703.1 CH940647 EDW68609.2 KQ971341 EFA12656.2 OUUW01000009 SPP85383.1 CH379068 EAL31218.1 CH479246 EDW37799.1 GEZM01021636 JAV88890.1 CH916366 EDV95795.1 KPJ13702.1 PCG78577.1 SPP85384.1 CH933816 EDW17351.2 GGFJ01007589 MBW56730.1 GGFJ01007588 MBW56729.1 GGFJ01007591 MBW56732.1 GFDL01005759 JAV29286.1 CH935273 EDW05269.2 GFDF01005883 JAV08201.1 GFDF01005884 JAV08200.1 UFQS01000006 UFQT01000006 SSW97057.1 SSX17444.1
RVE49426.1 NWSH01000191 PCG78578.1 KQ459602 KPI93450.1 KQ460615 KPJ13701.1 AGBW02011160 OWR47115.1 APCN01003099 AXCN02000230 AAAB01008844 EAA05949.4 CP012525 ALC44004.1 ALC43563.1 CM000159 EDW93072.1 CH954178 EDV50588.2 CH964168 EDW80141.2 JRES01000919 KNC27262.1 CH480815 EDW40812.1 CM000363 CM002912 EDX09805.1 KMY98558.1 AE014296 BT128694 AAN11985.1 AEE69542.1 BT001823 AAN71578.1 KA644894 AFP59523.1 GAPW01003265 JAC10333.1 GDHF01002050 JAI50264.1 GGFM01006158 MBW26909.1 CH902618 EDV41114.1 ATLV01024686 KE525357 KFB51877.1 CVRI01000062 CRL04205.1 GBXI01001903 JAD12389.1 BABH01030111 GAKP01008969 JAC49983.1 RVE49427.1 JXUM01052337 KQ561729 KXJ77703.1 CH940647 EDW68609.2 KQ971341 EFA12656.2 OUUW01000009 SPP85383.1 CH379068 EAL31218.1 CH479246 EDW37799.1 GEZM01021636 JAV88890.1 CH916366 EDV95795.1 KPJ13702.1 PCG78577.1 SPP85384.1 CH933816 EDW17351.2 GGFJ01007589 MBW56730.1 GGFJ01007588 MBW56729.1 GGFJ01007591 MBW56732.1 GFDL01005759 JAV29286.1 CH935273 EDW05269.2 GFDF01005883 JAV08201.1 GFDF01005884 JAV08200.1 UFQS01000006 UFQT01000006 SSW97057.1 SSX17444.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000007151 UP000075885 UP000075884 UP000075840 UP000075886 UP000075902 UP000075882 UP000007062 UP000076407 UP000095300 UP000092553 UP000075903 UP000002282 UP000008711 UP000007798 UP000076408 UP000192221 UP000075920 UP000075900 UP000037069 UP000001292 UP000075880 UP000000304 UP000000803 UP000095301 UP000007801 UP000030765 UP000183832 UP000091820 UP000069940 UP000249989 UP000008792 UP000007266 UP000268350 UP000001819 UP000008744 UP000001070 UP000009192 UP000078200 UP000069272
UP000007151 UP000075885 UP000075884 UP000075840 UP000075886 UP000075902 UP000075882 UP000007062 UP000076407 UP000095300 UP000092553 UP000075903 UP000002282 UP000008711 UP000007798 UP000076408 UP000192221 UP000075920 UP000075900 UP000037069 UP000001292 UP000075880 UP000000304 UP000000803 UP000095301 UP000007801 UP000030765 UP000183832 UP000091820 UP000069940 UP000249989 UP000008792 UP000007266 UP000268350 UP000001819 UP000008744 UP000001070 UP000009192 UP000078200 UP000069272
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9JDC8
A0A0L7L0K2
A0A2H1X164
A0A437BG64
A0A2A4K3U8
A0A194PQP4
+ More
A0A194RCR7 A0A212F032 A0A182PBA8 A0A182NSQ5 A0A182I8Q1 A0A182QT35 A0A182U7L9 A0A182LAL0 Q7QFU6 A0A182WV25 A0A1I8QBA5 A0A0M4E9Z5 A0A182URT5 A0A0M3QW56 B4PFE6 B3NFP6 B4N702 A0A182XWN7 A0A1W4V043 A0A182VY76 A0A182RDB8 A0A0L0C4T5 B4HKK9 A0A182JDM6 B4QMQ1 Q8IQB7 Q8IGE3 T1P8D8 A0A1I8MMX4 A0A023EM12 A0A0K8WGU4 A0A2M3ZEH9 B3M8W3 A0A084WNT3 A0A1J1IVG7 A0A1A9W7L7 A0A0A1XM50 H9JDC9 A0A034W876 A0A437BG67 A0A182GB94 B4LB45 D7EJD9 A0A3B0JWC0 Q29F54 B4HB05 A0A1Y1MWF7 B4J0X4 A0A194R7S7 A0A2A4K2V2 A0A3B0JTD7 B4L9H6 A0A2M4BUH0 A0A2M4BUJ8 A0A2M4BUQ9 A0A1Q3FP71 B4LAN8 A0A1A9VEA4 A0A1L8DP78 A0A182F7U6 A0A1L8DPE1 A0A336JY31
A0A194RCR7 A0A212F032 A0A182PBA8 A0A182NSQ5 A0A182I8Q1 A0A182QT35 A0A182U7L9 A0A182LAL0 Q7QFU6 A0A182WV25 A0A1I8QBA5 A0A0M4E9Z5 A0A182URT5 A0A0M3QW56 B4PFE6 B3NFP6 B4N702 A0A182XWN7 A0A1W4V043 A0A182VY76 A0A182RDB8 A0A0L0C4T5 B4HKK9 A0A182JDM6 B4QMQ1 Q8IQB7 Q8IGE3 T1P8D8 A0A1I8MMX4 A0A023EM12 A0A0K8WGU4 A0A2M3ZEH9 B3M8W3 A0A084WNT3 A0A1J1IVG7 A0A1A9W7L7 A0A0A1XM50 H9JDC9 A0A034W876 A0A437BG67 A0A182GB94 B4LB45 D7EJD9 A0A3B0JWC0 Q29F54 B4HB05 A0A1Y1MWF7 B4J0X4 A0A194R7S7 A0A2A4K2V2 A0A3B0JTD7 B4L9H6 A0A2M4BUH0 A0A2M4BUJ8 A0A2M4BUQ9 A0A1Q3FP71 B4LAN8 A0A1A9VEA4 A0A1L8DP78 A0A182F7U6 A0A1L8DPE1 A0A336JY31
Ontologies
GO
Topology
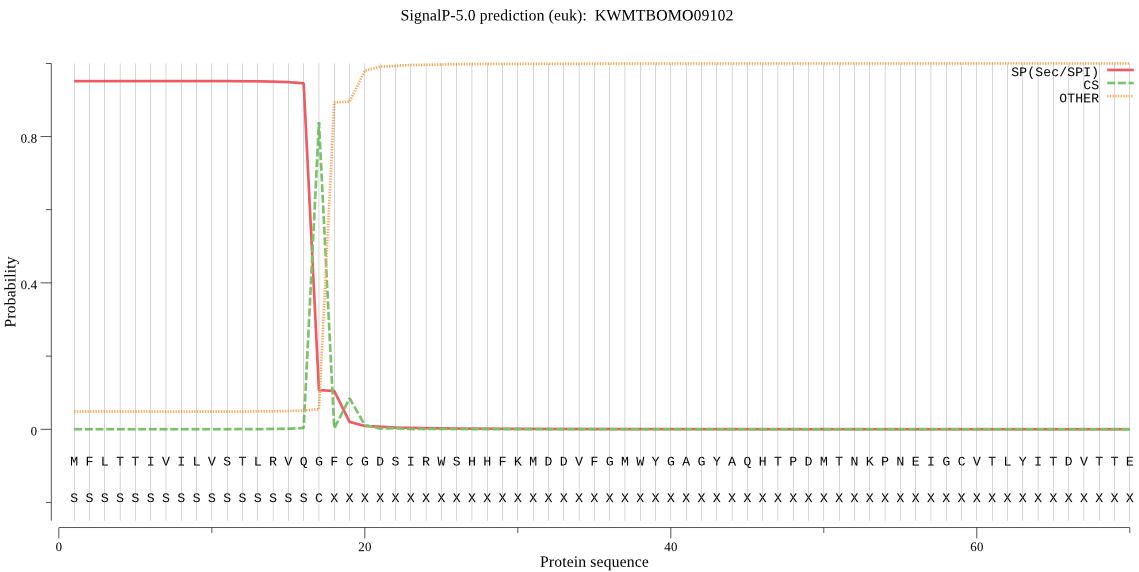
SignalP
Position: 1 - 17,
Likelihood: 0.950983
Length:
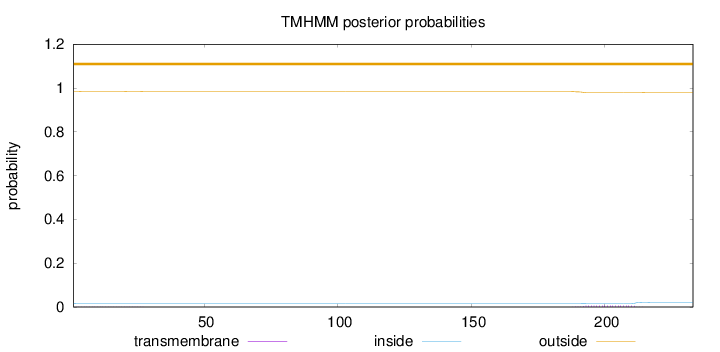
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14869
Exp number, first 60 AAs:
0.01595
Total prob of N-in:
0.01633
outside
1 - 233
Population Genetic Test Statistics
Pi
161.138019
Theta
143.205037
Tajima's D
0.347714
CLR
0.018469
CSRT
0.464376781160942
Interpretation
Uncertain
