Gene
KWMTBOMO09099
Pre Gene Modal
BGIBMGA007573
Annotation
PREDICTED:_uncharacterized_protein_LOC101742248_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.559
Sequence
CDS
ATGGCGCTTGTAATGCTGCCTTGCGACCTACCCTGGTGGACGTCTGTTCAACGACATCTCAAACATCTTCTGGTAGCTTCTTCGCCAACTAAGCTAACAGCAAGCATGTTGAAAATACACGACATGTGCAATATCGGGATAGATCCGGACGATGATATCAAGGATCCAGATCTAATGAAAGGCTTAGAGCAATTCCTCGAAGAGGAGATGAACGAAGAAGAACGGTCGCATTTCTTAGACAATACTGTTAGGATTATGGTGAATAGGGCACTGCACCTTAAAAGATGGCGCCCGCCAAAGGGACTGATGTTTAGCTTACAACAACAAAGTGACGTTACAGAACTAGATTATAACTTTGTATCGTCGCTAATCGCTCATGCCTTATTCTCCACGTTTCCGAAGAGAACGCTCAAGACTCATCCCACATTACAAGACTTTAATTTTACACATTTTTTTAAGAATTTACACAGAAAATCGCAACGGAACAAGTTAAAAAGTTTATTACATTACTTCGAGTGGTTGGACAAACACAATAATGAAGGGTCCATAAAAATTAGTCGGCAGGTAATGACATCGAAACAATGGTTGACCATAGAAGACTGGTTAGAATGCACGTTGCCGCTGTGCAAGCTGCTGGTTAGGCACGAGGGACGACCAGAAAGATGTGAGAACGAAGACGCCATTAGAGTGTGCTTCGCTACAAGCAGGATCGGGGGCAACATACTAATCGACGGAGAATCTCAGGAATCACTGTCGATGTTCATGATGCCAGAGCTATTGTCCACTATGCTATCAGTAGAATCTCTCGAAGATAATGAAGTTATTAAAGTGGAGGGAGCGAGAATGTTCAGCAGGATATACGATAGAAAACAAAAGGCTACGCTAGAACTACTCGAAGAACCTAAAAAAGTAACGGTGTGTCTGATGGATGCAGAAGACTACAGCAGTCTCCCGTTAGGTCAGTGGGAAGAGGACAATGTACTACGTGAACTAAACAAGAGTCTGCTTGCGTTCCAGCAAACACCGATGAAAAGTAAAGATACCATTCGTCACGAGAGACGCTTGTCGCCTATAGGCGAATCCTTTAGCCACACGCCGCCAGAAGTGGAAGCCACGGTGGTCATCAAACAGGCTTCGTCCTGCAGCACCATCAACACCAACAGCAGGAGCCCGTCGCCGCAGAACTACTGCGCAGCGACCCTCAACCTCAACGACCCCAGTGTGGAACTGTCTAAGCGTAAGTGCTGGCTCTCCCCGGACGGTGGGATGTTGAACAATCGCCGTGGACGATTCATAGTGCTCGGCTCGTCTGGGGAATGTTTGCCAGTGACCAGAAGCACGGGCAACGACCAAGACACGCAGGAGGACAGCTTGTATTCGAGCTGCAACTCCAGCGAAGACGAGTATCACAGTGCCAATGATAGTTTTGATTACGGTAGTGAAGACGAAGGCAGAGGAGAGAGCGCCCACCCAAATTCATTCCAGTATAACAAAGAATTATCAACCGAAGAGAAGAGGATTTCGTTTGCGGATAAACTCAAGGAGGCTTTGCGGAGAGAAGCTGAGGACTCTTGTACTAGTAGCAACACCGGGGACTGGTCCACGGACAGCTCTAGCTATGCTGTCGGCATTAGCATCTCGGGAGCTGGAATTAATGATAACGAAATAAGAATAAAGCGCGGCGGCTCAGGGGGCTTCGTGCTGACGGACAAGGAAACGACAGTCAACGGCGACTTGTCTCCGAGACGAAGACCTCTCGGGAGGAAGGACACGGGAAACAGTTCTAAGTACAGTTTCAGCACCGAATACACATCAGAATTGGAGGAGGTTTACGAGCAGTTCAACCAATGGCTGAACGATCCCATAATGGACGCGGGCTCTGGAGAACACAAAGCACGAGAATTGGACTCTCGCGACTTGGCTGTCATAAGGTACTTAATAAGTATTTTGGTAGCGCACCCCGACCTACCATAA
Protein
MALVMLPCDLPWWTSVQRHLKHLLVASSPTKLTASMLKIHDMCNIGIDPDDDIKDPDLMKGLEQFLEEEMNEEERSHFLDNTVRIMVNRALHLKRWRPPKGLMFSLQQQSDVTELDYNFVSSLIAHALFSTFPKRTLKTHPTLQDFNFTHFFKNLHRKSQRNKLKSLLHYFEWLDKHNNEGSIKISRQVMTSKQWLTIEDWLECTLPLCKLLVRHEGRPERCENEDAIRVCFATSRIGGNILIDGESQESLSMFMMPELLSTMLSVESLEDNEVIKVEGARMFSRIYDRKQKATLELLEEPKKVTVCLMDAEDYSSLPLGQWEEDNVLRELNKSLLAFQQTPMKSKDTIRHERRLSPIGESFSHTPPEVEATVVIKQASSCSTINTNSRSPSPQNYCAATLNLNDPSVELSKRKCWLSPDGGMLNNRRGRFIVLGSSGECLPVTRSTGNDQDTQEDSLYSSCNSSEDEYHSANDSFDYGSEDEGRGESAHPNSFQYNKELSTEEKRISFADKLKEALRREAEDSCTSSNTGDWSTDSSSYAVGISISGAGINDNEIRIKRGGSGGFVLTDKETTVNGDLSPRRRPLGRKDTGNSSKYSFSTEYTSELEEVYEQFNQWLNDPIMDAGSGEHKARELDSRDLAVIRYLISILVAHPDLP
Summary
Uniprot
H9JDH6
A0A2H1WXC8
A0A3S2M214
A0A212F048
A0A194R797
A0A194PJE6
+ More
A0A0L7LJB3 A0A1Y1M2C0 A0A336LIK3 A0A336LHU1 A0A0L7RIY4 A0A154NZP7 A0A1W4XW16 A0A1W4XV06 A0A310S421 A0A2A3EA87 A0A067R1U5 A0A088AHH1 A0A0T6AY66 A0A2J7R8B9 A0A3L8DNF3 A0A158NNS8 F4WV13 A0A026WGS2 E2A3E3 A0A182WV27 A0A1Y1M1G3 A0A182VY78 A0A084WNT4 A0A182KCP5 A0A182I8Q3 A0A1Y9IWK9 A0A0N0U2Y2 Q7QFU8 A0A182XWN5 A0A182RDB6 A0A151I739 A0A195BS23 A0A182PBB0 A0A1S4FH28 Q171S9 A0A195EX54 A0A232FE57 A0A151WSV9 A0A1Q3FIR3 A0A182GB96 B0WLZ8 A0A182GG47 A0A1Q3FYJ6 A0A151J1Y8 W5JCH6 A0A1B0CZ63 A0A182NSQ7 A0A182F7U8 A0A2J7R8C6 A0A1B6K6V4 A0A1B6HPD4 U4U8N4 A0A1W4XKW2 A0A1B6EEU9 A0A2J7R8B2 A0A182ING1 A0A182V304 A0A182LAK9 A0A182QAB6 A0A182MWI1 K7IVD7 A0A3Q0J9I2 A0A1B0GK92 A0A182SVF4 A0A1B6LBC6 A0A0A9YQ39 A0A146LEU2 A0A182U7A1 A0A023EX02 A0A2P8ZBX7 A0A1J1IB10 A0A0J7KJQ0 A0A0P4Y2P0 A0A0P5HVE1 A0A0P5DD43 A0A0P5GIY2 A0A0P5EIU2 A0A0P5LDP8 A0A0P5E656 A0A0N8AQL1 A0A0P5IL22 A0A0P5Y0H9 A0A0P5PH42 A0A0P5P8E1 A0A0P5GHU6 A0A0P5GAE0 A0A0N8DW47 A0A0P5GMH0 A0A0N8DAC0 A0A0P4YMZ6
A0A0L7LJB3 A0A1Y1M2C0 A0A336LIK3 A0A336LHU1 A0A0L7RIY4 A0A154NZP7 A0A1W4XW16 A0A1W4XV06 A0A310S421 A0A2A3EA87 A0A067R1U5 A0A088AHH1 A0A0T6AY66 A0A2J7R8B9 A0A3L8DNF3 A0A158NNS8 F4WV13 A0A026WGS2 E2A3E3 A0A182WV27 A0A1Y1M1G3 A0A182VY78 A0A084WNT4 A0A182KCP5 A0A182I8Q3 A0A1Y9IWK9 A0A0N0U2Y2 Q7QFU8 A0A182XWN5 A0A182RDB6 A0A151I739 A0A195BS23 A0A182PBB0 A0A1S4FH28 Q171S9 A0A195EX54 A0A232FE57 A0A151WSV9 A0A1Q3FIR3 A0A182GB96 B0WLZ8 A0A182GG47 A0A1Q3FYJ6 A0A151J1Y8 W5JCH6 A0A1B0CZ63 A0A182NSQ7 A0A182F7U8 A0A2J7R8C6 A0A1B6K6V4 A0A1B6HPD4 U4U8N4 A0A1W4XKW2 A0A1B6EEU9 A0A2J7R8B2 A0A182ING1 A0A182V304 A0A182LAK9 A0A182QAB6 A0A182MWI1 K7IVD7 A0A3Q0J9I2 A0A1B0GK92 A0A182SVF4 A0A1B6LBC6 A0A0A9YQ39 A0A146LEU2 A0A182U7A1 A0A023EX02 A0A2P8ZBX7 A0A1J1IB10 A0A0J7KJQ0 A0A0P4Y2P0 A0A0P5HVE1 A0A0P5DD43 A0A0P5GIY2 A0A0P5EIU2 A0A0P5LDP8 A0A0P5E656 A0A0N8AQL1 A0A0P5IL22 A0A0P5Y0H9 A0A0P5PH42 A0A0P5P8E1 A0A0P5GHU6 A0A0P5GAE0 A0A0N8DW47 A0A0P5GMH0 A0A0N8DAC0 A0A0P4YMZ6
Pubmed
EMBL
BABH01030112
ODYU01011781
SOQ57720.1
RSAL01000065
RVE49423.1
AGBW02011160
+ More
OWR47119.1 KQ460615 KPJ13698.1 KQ459602 KPI93447.1 JTDY01000861 KOB75638.1 GEZM01042796 GEZM01042795 JAV79591.1 UFQS01000006 UFQT01000006 SSW97058.1 SSX17445.1 SSW97059.1 SSX17446.1 KQ414583 KOC70768.1 KQ434786 KZC05097.1 KQ794714 OAD47011.1 KZ288310 PBC28628.1 KK852767 KDR16965.1 LJIG01022557 KRT79940.1 NEVH01006723 PNF37077.1 QOIP01000006 RLU21970.1 ADTU01021750 ADTU01021751 ADTU01021752 GL888384 EGI61901.1 KK107256 EZA54279.1 GL436428 EFN72007.1 GEZM01042800 GEZM01042799 JAV79583.1 ATLV01024689 KE525357 KFB51878.1 APCN01003099 KQ435969 KOX67874.1 AAAB01008844 EAA05953.4 KQ978438 KYM93982.1 KQ976417 KYM89815.1 CH477447 EAT40738.1 KQ981953 KYN32482.1 NNAY01000354 OXU28965.1 KQ982766 KYQ50989.1 GFDL01007534 JAV27511.1 JXUM01052341 JXUM01052342 JXUM01052343 JXUM01052344 KQ561729 KXJ77705.1 DS231993 EDS30812.1 JXUM01061296 JXUM01061297 JXUM01061298 JXUM01061299 JXUM01061300 KQ562144 KXJ76581.1 GFDL01002390 JAV32655.1 KQ980503 KYN15793.1 ADMH02001894 ETN60585.1 AJVK01020640 PNF37079.1 GECU01000526 JAT07181.1 GECU01031205 JAS76501.1 KB631899 ERL86956.1 GEDC01020773 GEDC01000832 JAS16525.1 JAS36466.1 PNF37076.1 AXCN02000230 AXCM01009907 AAZX01005070 AAZX01005358 AJWK01028557 AJWK01028558 AJWK01028559 AJWK01028560 GEBQ01019036 JAT20941.1 GBHO01009848 GBHO01009752 JAG33756.1 JAG33852.1 GDHC01011846 GDHC01010215 JAQ06783.1 JAQ08414.1 GBBI01004807 JAC13905.1 PYGN01000110 PSN54006.1 CVRI01000047 CRK97424.1 LBMM01006608 KMQ90466.1 GDIP01233407 JAI89994.1 GDIQ01224590 JAK27135.1 GDIP01158063 JAJ65339.1 GDIQ01243973 JAK07752.1 GDIP01145878 JAJ77524.1 GDIQ01171191 JAK80534.1 GDIP01152275 JAJ71127.1 GDIQ01252571 JAJ99153.1 GDIQ01214183 JAK37542.1 GDIP01064733 JAM38982.1 GDIQ01130968 JAL20758.1 GDIQ01132273 JAL19453.1 GDIQ01242276 JAK09449.1 GDIQ01243972 JAK07753.1 GDIQ01086092 JAN08645.1 GDIQ01239626 JAK12099.1 GDIP01053287 JAM50428.1 GDIP01225159 JAI98242.1
OWR47119.1 KQ460615 KPJ13698.1 KQ459602 KPI93447.1 JTDY01000861 KOB75638.1 GEZM01042796 GEZM01042795 JAV79591.1 UFQS01000006 UFQT01000006 SSW97058.1 SSX17445.1 SSW97059.1 SSX17446.1 KQ414583 KOC70768.1 KQ434786 KZC05097.1 KQ794714 OAD47011.1 KZ288310 PBC28628.1 KK852767 KDR16965.1 LJIG01022557 KRT79940.1 NEVH01006723 PNF37077.1 QOIP01000006 RLU21970.1 ADTU01021750 ADTU01021751 ADTU01021752 GL888384 EGI61901.1 KK107256 EZA54279.1 GL436428 EFN72007.1 GEZM01042800 GEZM01042799 JAV79583.1 ATLV01024689 KE525357 KFB51878.1 APCN01003099 KQ435969 KOX67874.1 AAAB01008844 EAA05953.4 KQ978438 KYM93982.1 KQ976417 KYM89815.1 CH477447 EAT40738.1 KQ981953 KYN32482.1 NNAY01000354 OXU28965.1 KQ982766 KYQ50989.1 GFDL01007534 JAV27511.1 JXUM01052341 JXUM01052342 JXUM01052343 JXUM01052344 KQ561729 KXJ77705.1 DS231993 EDS30812.1 JXUM01061296 JXUM01061297 JXUM01061298 JXUM01061299 JXUM01061300 KQ562144 KXJ76581.1 GFDL01002390 JAV32655.1 KQ980503 KYN15793.1 ADMH02001894 ETN60585.1 AJVK01020640 PNF37079.1 GECU01000526 JAT07181.1 GECU01031205 JAS76501.1 KB631899 ERL86956.1 GEDC01020773 GEDC01000832 JAS16525.1 JAS36466.1 PNF37076.1 AXCN02000230 AXCM01009907 AAZX01005070 AAZX01005358 AJWK01028557 AJWK01028558 AJWK01028559 AJWK01028560 GEBQ01019036 JAT20941.1 GBHO01009848 GBHO01009752 JAG33756.1 JAG33852.1 GDHC01011846 GDHC01010215 JAQ06783.1 JAQ08414.1 GBBI01004807 JAC13905.1 PYGN01000110 PSN54006.1 CVRI01000047 CRK97424.1 LBMM01006608 KMQ90466.1 GDIP01233407 JAI89994.1 GDIQ01224590 JAK27135.1 GDIP01158063 JAJ65339.1 GDIQ01243973 JAK07752.1 GDIP01145878 JAJ77524.1 GDIQ01171191 JAK80534.1 GDIP01152275 JAJ71127.1 GDIQ01252571 JAJ99153.1 GDIQ01214183 JAK37542.1 GDIP01064733 JAM38982.1 GDIQ01130968 JAL20758.1 GDIQ01132273 JAL19453.1 GDIQ01242276 JAK09449.1 GDIQ01243972 JAK07753.1 GDIQ01086092 JAN08645.1 GDIQ01239626 JAK12099.1 GDIP01053287 JAM50428.1 GDIP01225159 JAI98242.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000053825 UP000076502 UP000192223 UP000242457 UP000027135 UP000005203 UP000235965 UP000279307 UP000005205 UP000007755 UP000053097 UP000000311 UP000076407 UP000075920 UP000030765 UP000075881 UP000075840 UP000053105 UP000007062 UP000076408 UP000075900 UP000078542 UP000078540 UP000075885 UP000008820 UP000078541 UP000215335 UP000075809 UP000069940 UP000249989 UP000002320 UP000078492 UP000000673 UP000092462 UP000075884 UP000069272 UP000030742 UP000075880 UP000075903 UP000075882 UP000075886 UP000075883 UP000002358 UP000079169 UP000092461 UP000075901 UP000075902 UP000245037 UP000183832 UP000036403
UP000053825 UP000076502 UP000192223 UP000242457 UP000027135 UP000005203 UP000235965 UP000279307 UP000005205 UP000007755 UP000053097 UP000000311 UP000076407 UP000075920 UP000030765 UP000075881 UP000075840 UP000053105 UP000007062 UP000076408 UP000075900 UP000078542 UP000078540 UP000075885 UP000008820 UP000078541 UP000215335 UP000075809 UP000069940 UP000249989 UP000002320 UP000078492 UP000000673 UP000092462 UP000075884 UP000069272 UP000030742 UP000075880 UP000075903 UP000075882 UP000075886 UP000075883 UP000002358 UP000079169 UP000092461 UP000075901 UP000075902 UP000245037 UP000183832 UP000036403
PRIDE
Pfam
PF05028 PARG_cat
Interpro
IPR007724
Poly_GlycHdrlase
ProteinModelPortal
H9JDH6
A0A2H1WXC8
A0A3S2M214
A0A212F048
A0A194R797
A0A194PJE6
+ More
A0A0L7LJB3 A0A1Y1M2C0 A0A336LIK3 A0A336LHU1 A0A0L7RIY4 A0A154NZP7 A0A1W4XW16 A0A1W4XV06 A0A310S421 A0A2A3EA87 A0A067R1U5 A0A088AHH1 A0A0T6AY66 A0A2J7R8B9 A0A3L8DNF3 A0A158NNS8 F4WV13 A0A026WGS2 E2A3E3 A0A182WV27 A0A1Y1M1G3 A0A182VY78 A0A084WNT4 A0A182KCP5 A0A182I8Q3 A0A1Y9IWK9 A0A0N0U2Y2 Q7QFU8 A0A182XWN5 A0A182RDB6 A0A151I739 A0A195BS23 A0A182PBB0 A0A1S4FH28 Q171S9 A0A195EX54 A0A232FE57 A0A151WSV9 A0A1Q3FIR3 A0A182GB96 B0WLZ8 A0A182GG47 A0A1Q3FYJ6 A0A151J1Y8 W5JCH6 A0A1B0CZ63 A0A182NSQ7 A0A182F7U8 A0A2J7R8C6 A0A1B6K6V4 A0A1B6HPD4 U4U8N4 A0A1W4XKW2 A0A1B6EEU9 A0A2J7R8B2 A0A182ING1 A0A182V304 A0A182LAK9 A0A182QAB6 A0A182MWI1 K7IVD7 A0A3Q0J9I2 A0A1B0GK92 A0A182SVF4 A0A1B6LBC6 A0A0A9YQ39 A0A146LEU2 A0A182U7A1 A0A023EX02 A0A2P8ZBX7 A0A1J1IB10 A0A0J7KJQ0 A0A0P4Y2P0 A0A0P5HVE1 A0A0P5DD43 A0A0P5GIY2 A0A0P5EIU2 A0A0P5LDP8 A0A0P5E656 A0A0N8AQL1 A0A0P5IL22 A0A0P5Y0H9 A0A0P5PH42 A0A0P5P8E1 A0A0P5GHU6 A0A0P5GAE0 A0A0N8DW47 A0A0P5GMH0 A0A0N8DAC0 A0A0P4YMZ6
A0A0L7LJB3 A0A1Y1M2C0 A0A336LIK3 A0A336LHU1 A0A0L7RIY4 A0A154NZP7 A0A1W4XW16 A0A1W4XV06 A0A310S421 A0A2A3EA87 A0A067R1U5 A0A088AHH1 A0A0T6AY66 A0A2J7R8B9 A0A3L8DNF3 A0A158NNS8 F4WV13 A0A026WGS2 E2A3E3 A0A182WV27 A0A1Y1M1G3 A0A182VY78 A0A084WNT4 A0A182KCP5 A0A182I8Q3 A0A1Y9IWK9 A0A0N0U2Y2 Q7QFU8 A0A182XWN5 A0A182RDB6 A0A151I739 A0A195BS23 A0A182PBB0 A0A1S4FH28 Q171S9 A0A195EX54 A0A232FE57 A0A151WSV9 A0A1Q3FIR3 A0A182GB96 B0WLZ8 A0A182GG47 A0A1Q3FYJ6 A0A151J1Y8 W5JCH6 A0A1B0CZ63 A0A182NSQ7 A0A182F7U8 A0A2J7R8C6 A0A1B6K6V4 A0A1B6HPD4 U4U8N4 A0A1W4XKW2 A0A1B6EEU9 A0A2J7R8B2 A0A182ING1 A0A182V304 A0A182LAK9 A0A182QAB6 A0A182MWI1 K7IVD7 A0A3Q0J9I2 A0A1B0GK92 A0A182SVF4 A0A1B6LBC6 A0A0A9YQ39 A0A146LEU2 A0A182U7A1 A0A023EX02 A0A2P8ZBX7 A0A1J1IB10 A0A0J7KJQ0 A0A0P4Y2P0 A0A0P5HVE1 A0A0P5DD43 A0A0P5GIY2 A0A0P5EIU2 A0A0P5LDP8 A0A0P5E656 A0A0N8AQL1 A0A0P5IL22 A0A0P5Y0H9 A0A0P5PH42 A0A0P5P8E1 A0A0P5GHU6 A0A0P5GAE0 A0A0N8DW47 A0A0P5GMH0 A0A0N8DAC0 A0A0P4YMZ6
PDB
5A7R
E-value=4.54318e-15,
Score=200
Ontologies
GO
PANTHER
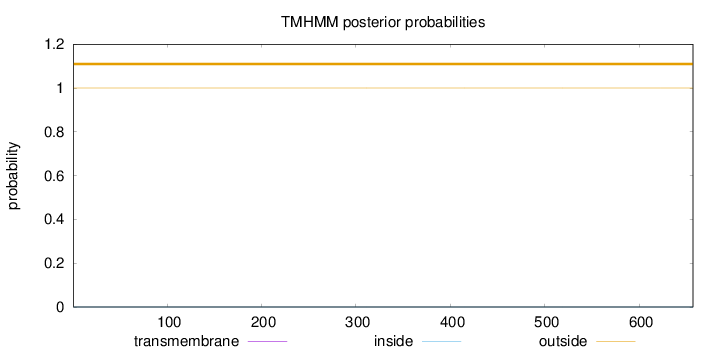
Topology
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00114
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00008
outside
1 - 657
Population Genetic Test Statistics
Pi
282.339569
Theta
200.776939
Tajima's D
1.086441
CLR
0.265202
CSRT
0.684315784210789
Interpretation
Uncertain
