Pre Gene Modal
BGIBMGA007515
Annotation
PREDICTED:_GRIP_and_coiled-coil_domain-containing_protein_2-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.302
Sequence
CDS
ATGGGTTCAGAGTCAGCGCCGTCTCCTCCACCATCTAAACCGTTCACGGGCGGGGGCAGTGGTCGCTCCCCGGTCCCTCTGGAGCGACTGCTCGAGGAAGGAGTGCCCGATGATGAAGCGCTCGACACGTCTTCATTAGGCTTCACCCCAGAACAGGAGGTGGCCGATCTAAAAAGGCGTTTACAAGCTCAACAACAAAGAGTGAAGCACGTCACGGTGCTGCTATCGGAGTCTGAGCGAGAATGCGCGCGTATGGCTCAGCTCAGCGAACTCCTTAAGGCAGAGTTGCGTCGGGTGAGAGGGACCACGCAAAATGCACATAACACCGAGTACATGAAGAACGTCACTCTCAAGTTCCTAACCCTTCCATCCGGTGACGAGCGTATTCGACTTGTCCCAGTTCTTCAGAAGATTTTGACCTTGACGCCAGACGAAACTCAAAAAATTCAAGCGATTGCAAAAGGTTTAGATCCAAACTCGAGTAAAGGATGGGGAAGCTACTTGCCGTGGCCTGGAGGGAAATGA
Protein
MGSESAPSPPPSKPFTGGGSGRSPVPLERLLEEGVPDDEALDTSSLGFTPEQEVADLKRRLQAQQQRVKHVTVLLSESERECARMAQLSELLKAELRRVRGTTQNAHNTEYMKNVTLKFLTLPSGDERIRLVPVLQKILTLTPDETQKIQAIAKGLDPNSSKGWGSYLPWPGGK
Summary
Uniprot
H9JDB8
A0A2A4JZI8
A0A194PL30
A0A437BL54
A0A212EL07
A0A0K8TUK9
+ More
W5JIZ8 A0A2M3ZJY1 D6WKQ2 A0A067R2L1 A0A182K560 A0A182F7Q9 A0A2M4A721 A0A2M4A637 A0A2J7QL96 A0A1A9VBD0 A0A1A9YG62 A0A1B0BT67 A0A1L8DW13 A0A2P8YTZ9 A0A182VFI1 A0A182UHY4 A0A1Q3FK61 Q5TT60 A0A182WVW7 A0A182HZ71 A0A1B0F9V4 A0A2J7QLC2 A0A182KXD9 A0A182R8C3 U4USN3 A0A182P0B9 A0A2J7QLA2 A0A2J7QL95 A0A1A9ZNU9 A0A182NSM2 A0A182MVY5 A0A182VS19 Q176W4 A0A1S4FD08 A0A1I8M2F1 A0A1A9WJE0 A0A1B6GJD1 A0A0T6AXM3 A0A1I8Q1P1 E0VFU3 A0A0K8T0I1 A0A084WRU9 A0A0A9YCF3 A0A1B6DFM6 A0A182GHS0 A0A0A9YJZ6 A0A336KMP6 A0A444TAY4 A0A1B6LE98 A0A2S2R5G5 R7UX32 B3P1H6 B4K5L7 A0A1Y1K9N7 A0A1Y1KEX4 A0A3B0K8Q0 A0A3B0KDX5 A0A3B0KL03 A0A0K8R2T9 A0A224XHK2 Q32KE8 Q9VGE4 A0A3Q0J439 A0A087U7U8 B3M033 A0A069DYE9 B4PN19 A0A0Q9X4H9 A0A1D2MV73 A0A1S3K777 A0A1W4UMQ1 V5GWQ1 T1J702 A0A2P2HYK8 B4JT77 W4XJE3 B4QTI0 B4HHI3
W5JIZ8 A0A2M3ZJY1 D6WKQ2 A0A067R2L1 A0A182K560 A0A182F7Q9 A0A2M4A721 A0A2M4A637 A0A2J7QL96 A0A1A9VBD0 A0A1A9YG62 A0A1B0BT67 A0A1L8DW13 A0A2P8YTZ9 A0A182VFI1 A0A182UHY4 A0A1Q3FK61 Q5TT60 A0A182WVW7 A0A182HZ71 A0A1B0F9V4 A0A2J7QLC2 A0A182KXD9 A0A182R8C3 U4USN3 A0A182P0B9 A0A2J7QLA2 A0A2J7QL95 A0A1A9ZNU9 A0A182NSM2 A0A182MVY5 A0A182VS19 Q176W4 A0A1S4FD08 A0A1I8M2F1 A0A1A9WJE0 A0A1B6GJD1 A0A0T6AXM3 A0A1I8Q1P1 E0VFU3 A0A0K8T0I1 A0A084WRU9 A0A0A9YCF3 A0A1B6DFM6 A0A182GHS0 A0A0A9YJZ6 A0A336KMP6 A0A444TAY4 A0A1B6LE98 A0A2S2R5G5 R7UX32 B3P1H6 B4K5L7 A0A1Y1K9N7 A0A1Y1KEX4 A0A3B0K8Q0 A0A3B0KDX5 A0A3B0KL03 A0A0K8R2T9 A0A224XHK2 Q32KE8 Q9VGE4 A0A3Q0J439 A0A087U7U8 B3M033 A0A069DYE9 B4PN19 A0A0Q9X4H9 A0A1D2MV73 A0A1S3K777 A0A1W4UMQ1 V5GWQ1 T1J702 A0A2P2HYK8 B4JT77 W4XJE3 B4QTI0 B4HHI3
Pubmed
19121390
26354079
22118469
26369729
20920257
23761445
+ More
18362917 19820115 24845553 29403074 12364791 14747013 17210077 20966253 23537049 17510324 25315136 20566863 24438588 25401762 26483478 23254933 17994087 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 17550304 27289101 25765539
18362917 19820115 24845553 29403074 12364791 14747013 17210077 20966253 23537049 17510324 25315136 20566863 24438588 25401762 26483478 23254933 17994087 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 17550304 27289101 25765539
EMBL
BABH01030128
BABH01030129
BABH01030130
BABH01030131
BABH01030132
BABH01030133
+ More
NWSH01000353 PCG77098.1 KQ459602 KPI93434.1 RSAL01000040 RVE50995.1 AGBW02014166 OWR42157.1 GDAI01000003 JAI17600.1 ADMH02001358 ETN62860.1 GGFM01008034 MBW28785.1 KQ971342 EFA03027.1 KK852751 KDR17156.1 GGFK01003107 MBW36428.1 GGFK01002946 MBW36267.1 NEVH01013250 PNF29355.1 JXJN01020019 GFDF01003453 JAV10631.1 PYGN01000359 PSN47728.1 GFDL01007055 JAV27990.1 AAAB01008888 EAL40582.4 APCN01005504 CCAG010002599 PNF29358.1 KB632348 ERL93186.1 PNF29357.1 PNF29356.1 AXCM01000837 CH477381 EAT42208.1 GECZ01007376 JAS62393.1 LJIG01022555 KRT79952.1 DS235124 EEB12249.1 GBRD01007116 JAG58705.1 ATLV01026212 ATLV01026213 ATLV01026214 ATLV01026215 KE525408 KFB52943.1 GBHO01013760 GBRD01007115 JAG29844.1 JAG58706.1 GEDC01012876 JAS24422.1 JXUM01064417 KQ562297 KXJ76211.1 GBHO01013759 JAG29845.1 UFQS01000679 UFQT01000679 SSX06136.1 SSX26492.1 SAUD01005646 RXG67132.1 GEBQ01017960 JAT22017.1 GGMS01016053 MBY85256.1 AMQN01005951 KB297182 ELU10899.1 CH954181 EDV49435.1 CH933806 EDW14054.1 GEZM01088477 JAV58179.1 GEZM01088478 JAV58175.1 OUUW01000008 SPP84480.1 SPP84479.1 SPP84478.1 GADI01008428 JAA65380.1 GFTR01008813 JAW07613.1 BT023931 ABB36435.1 AE014297 BT046132 AAF54739.3 ACI46520.1 KK118615 KFM73437.1 CH902617 EDV41990.1 GBGD01000077 JAC88812.1 CM000160 EDW97031.1 KRG00826.1 LJIJ01000479 ODM96997.1 GANP01009623 JAB74845.1 JH431901 IACF01001151 LAB66868.1 CH916373 EDV94967.1 AAGJ04051875 CM000364 EDX13285.1 CH480815 EDW42522.1
NWSH01000353 PCG77098.1 KQ459602 KPI93434.1 RSAL01000040 RVE50995.1 AGBW02014166 OWR42157.1 GDAI01000003 JAI17600.1 ADMH02001358 ETN62860.1 GGFM01008034 MBW28785.1 KQ971342 EFA03027.1 KK852751 KDR17156.1 GGFK01003107 MBW36428.1 GGFK01002946 MBW36267.1 NEVH01013250 PNF29355.1 JXJN01020019 GFDF01003453 JAV10631.1 PYGN01000359 PSN47728.1 GFDL01007055 JAV27990.1 AAAB01008888 EAL40582.4 APCN01005504 CCAG010002599 PNF29358.1 KB632348 ERL93186.1 PNF29357.1 PNF29356.1 AXCM01000837 CH477381 EAT42208.1 GECZ01007376 JAS62393.1 LJIG01022555 KRT79952.1 DS235124 EEB12249.1 GBRD01007116 JAG58705.1 ATLV01026212 ATLV01026213 ATLV01026214 ATLV01026215 KE525408 KFB52943.1 GBHO01013760 GBRD01007115 JAG29844.1 JAG58706.1 GEDC01012876 JAS24422.1 JXUM01064417 KQ562297 KXJ76211.1 GBHO01013759 JAG29845.1 UFQS01000679 UFQT01000679 SSX06136.1 SSX26492.1 SAUD01005646 RXG67132.1 GEBQ01017960 JAT22017.1 GGMS01016053 MBY85256.1 AMQN01005951 KB297182 ELU10899.1 CH954181 EDV49435.1 CH933806 EDW14054.1 GEZM01088477 JAV58179.1 GEZM01088478 JAV58175.1 OUUW01000008 SPP84480.1 SPP84479.1 SPP84478.1 GADI01008428 JAA65380.1 GFTR01008813 JAW07613.1 BT023931 ABB36435.1 AE014297 BT046132 AAF54739.3 ACI46520.1 KK118615 KFM73437.1 CH902617 EDV41990.1 GBGD01000077 JAC88812.1 CM000160 EDW97031.1 KRG00826.1 LJIJ01000479 ODM96997.1 GANP01009623 JAB74845.1 JH431901 IACF01001151 LAB66868.1 CH916373 EDV94967.1 AAGJ04051875 CM000364 EDX13285.1 CH480815 EDW42522.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000000673
+ More
UP000007266 UP000027135 UP000075881 UP000069272 UP000235965 UP000078200 UP000092443 UP000092460 UP000245037 UP000075903 UP000075902 UP000007062 UP000076407 UP000075840 UP000092444 UP000075882 UP000075900 UP000030742 UP000075885 UP000092445 UP000075884 UP000075883 UP000075920 UP000008820 UP000095301 UP000091820 UP000095300 UP000009046 UP000030765 UP000069940 UP000249989 UP000288706 UP000014760 UP000008711 UP000009192 UP000268350 UP000000803 UP000079169 UP000054359 UP000007801 UP000002282 UP000094527 UP000085678 UP000192221 UP000001070 UP000007110 UP000000304 UP000001292
UP000007266 UP000027135 UP000075881 UP000069272 UP000235965 UP000078200 UP000092443 UP000092460 UP000245037 UP000075903 UP000075902 UP000007062 UP000076407 UP000075840 UP000092444 UP000075882 UP000075900 UP000030742 UP000075885 UP000092445 UP000075884 UP000075883 UP000075920 UP000008820 UP000095301 UP000091820 UP000095300 UP000009046 UP000030765 UP000069940 UP000249989 UP000288706 UP000014760 UP000008711 UP000009192 UP000268350 UP000000803 UP000079169 UP000054359 UP000007801 UP000002282 UP000094527 UP000085678 UP000192221 UP000001070 UP000007110 UP000000304 UP000001292
ProteinModelPortal
H9JDB8
A0A2A4JZI8
A0A194PL30
A0A437BL54
A0A212EL07
A0A0K8TUK9
+ More
W5JIZ8 A0A2M3ZJY1 D6WKQ2 A0A067R2L1 A0A182K560 A0A182F7Q9 A0A2M4A721 A0A2M4A637 A0A2J7QL96 A0A1A9VBD0 A0A1A9YG62 A0A1B0BT67 A0A1L8DW13 A0A2P8YTZ9 A0A182VFI1 A0A182UHY4 A0A1Q3FK61 Q5TT60 A0A182WVW7 A0A182HZ71 A0A1B0F9V4 A0A2J7QLC2 A0A182KXD9 A0A182R8C3 U4USN3 A0A182P0B9 A0A2J7QLA2 A0A2J7QL95 A0A1A9ZNU9 A0A182NSM2 A0A182MVY5 A0A182VS19 Q176W4 A0A1S4FD08 A0A1I8M2F1 A0A1A9WJE0 A0A1B6GJD1 A0A0T6AXM3 A0A1I8Q1P1 E0VFU3 A0A0K8T0I1 A0A084WRU9 A0A0A9YCF3 A0A1B6DFM6 A0A182GHS0 A0A0A9YJZ6 A0A336KMP6 A0A444TAY4 A0A1B6LE98 A0A2S2R5G5 R7UX32 B3P1H6 B4K5L7 A0A1Y1K9N7 A0A1Y1KEX4 A0A3B0K8Q0 A0A3B0KDX5 A0A3B0KL03 A0A0K8R2T9 A0A224XHK2 Q32KE8 Q9VGE4 A0A3Q0J439 A0A087U7U8 B3M033 A0A069DYE9 B4PN19 A0A0Q9X4H9 A0A1D2MV73 A0A1S3K777 A0A1W4UMQ1 V5GWQ1 T1J702 A0A2P2HYK8 B4JT77 W4XJE3 B4QTI0 B4HHI3
W5JIZ8 A0A2M3ZJY1 D6WKQ2 A0A067R2L1 A0A182K560 A0A182F7Q9 A0A2M4A721 A0A2M4A637 A0A2J7QL96 A0A1A9VBD0 A0A1A9YG62 A0A1B0BT67 A0A1L8DW13 A0A2P8YTZ9 A0A182VFI1 A0A182UHY4 A0A1Q3FK61 Q5TT60 A0A182WVW7 A0A182HZ71 A0A1B0F9V4 A0A2J7QLC2 A0A182KXD9 A0A182R8C3 U4USN3 A0A182P0B9 A0A2J7QLA2 A0A2J7QL95 A0A1A9ZNU9 A0A182NSM2 A0A182MVY5 A0A182VS19 Q176W4 A0A1S4FD08 A0A1I8M2F1 A0A1A9WJE0 A0A1B6GJD1 A0A0T6AXM3 A0A1I8Q1P1 E0VFU3 A0A0K8T0I1 A0A084WRU9 A0A0A9YCF3 A0A1B6DFM6 A0A182GHS0 A0A0A9YJZ6 A0A336KMP6 A0A444TAY4 A0A1B6LE98 A0A2S2R5G5 R7UX32 B3P1H6 B4K5L7 A0A1Y1K9N7 A0A1Y1KEX4 A0A3B0K8Q0 A0A3B0KDX5 A0A3B0KL03 A0A0K8R2T9 A0A224XHK2 Q32KE8 Q9VGE4 A0A3Q0J439 A0A087U7U8 B3M033 A0A069DYE9 B4PN19 A0A0Q9X4H9 A0A1D2MV73 A0A1S3K777 A0A1W4UMQ1 V5GWQ1 T1J702 A0A2P2HYK8 B4JT77 W4XJE3 B4QTI0 B4HHI3
Ontologies
GO
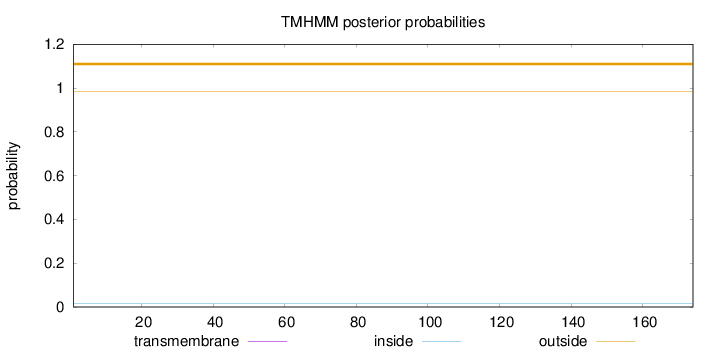
Topology
Length:
174
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01626
outside
1 - 174
Population Genetic Test Statistics
Pi
277.666416
Theta
206.972428
Tajima's D
1.180587
CLR
0
CSRT
0.702364881755912
Interpretation
Uncertain
