Gene
KWMTBOMO09079
Pre Gene Modal
BGIBMGA007579
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_transcription_initiation_factor_TFIID_subunit_6-like_[Bombyx_mori]
Full name
Transcription initiation factor TFIID subunit 6
Alternative Name
TAFII-60
TAFII-62
Transcription initiation factor TFIID 62 kDa subunit
TAFII-62
Transcription initiation factor TFIID 62 kDa subunit
Location in the cell
Nuclear Reliability : 3.151
Sequence
CDS
ATGGGCGACTCAGAATTGGCTTATGGATCTTCGTTAACGTTGGATTCCATGAAAGTGATAGCAGAGAGTGTTGGTATCGCCACTCTTGGCGATGACGCTGCTAAGGAACTCGCTGATGATGTTACATATCGCTTGAAAGTGATTGTTCAAGATGCCATGAAGTTCATGCATCACTCAAAGAGACAAAAACTTTCCATTACAGATATAGATCATGCCCTGAAGATAAAAAACAATGAGTGTCAATATGGATTTATTCAGCCTGATTCGCTGCCATTTAGGTTCGCATCTGGAGGAGGTAGAGAACTCCATTTCATTGAAGAAAAAGAGATAGATCTTTCGGAGATCCTATCAGCACCACCCCCAAAGGTTCCATTGGATGTATCTGTTCGTGCACATTGGCTAAGTGTGGATGGGGTTCAACCAACAGTACCTGAAAATCCACCACCGCTTTCAAAAGAAGCTCAGAAACTTGAATCTGTTGATCCAATAAGTAAACTGAGTAAACCAAGCAATAAAGATTCTGCAGGTAAACCTATATGTGGCAAAGCAGCAAGATTGAAAGCATCAGAATCTGTGCATGTTAAACAGCTTGCTACCCATGAATTGAGTGTTGAACAGCAGCTCTATTATAAAGAAATCACAGAAGCCTGTGTTGGCAGTGATGAAACAAGACGTGCTGAAGCGCTCCAGTCCTTAGCTTGTGACCCAGGACTACACGAGATGTTGCCCAGAATGTGTACTTTTATATCTGAAGGTGTGAAGGTTAATGTCGTACAAAACAATCTGGCTCTTCTCATTTACCTGATGCGAATGGTTAAGGCACTACTTGATAACCAGTCACTATATTTGGAAAAATATTTACACGAGTTGATTCCGTCCGTGTCGACATGCATAGTATCACGACAGCTGTGCCTTCGTCCCGAGATGGATAACCACTGGGCCCTGCGAGATTTCGCCGCGCGACTCATGGCTCAGATATGCAAAACATTCAATACTTCAACCAATAATCTACAAACGAGAGTCACCAGGTTGTTCGCGAAAGCCCTCCAGTGCCCTTCACAAACAAACAACGAAACGGGTCCTTCTATGGCGAGCATGAAGGAATCGGAGAAGACTCCTCTCGCGTCGCTGTATGGAGCCGTGCAAGGATTGGCAGAGCTGGGCCCCGAAGTCGTTAAAGTGTTCATTCTGCCGCGCGTGCGTTGGTTGGGAGAGCGCGTGGAGGGCGCGCTGAGCGGTATCGGTGGCGCGGACCGACTGGCGGCCGGCAACCTCAAGCACCAGCTGCTGAAAGTCCTGGCCCCTGTCGTCCGACAACTGCGGCAACCGCCCGACCAACCCGAAGACTACAAGCGCGACTTCGGGTACCTGGGTCCTAGTCTCCAACAAGCCGTCAGCAAGATACGCTCGTCTCCCTCGTGCAGTACCGCAGGAGGCGCCGTGGCGGTAGTCACCTGCACGCCGCCCCTATTGCCTACCGCGCCCTCGCCTTCGCCTTCCAATATATCGCATACTACAAAGACTGAAACAGTGACCACTCGTAACCTGGTGATAGCGTCATCGACGCCACAATCACCGGCCCCGTCTACACCGCCGCCACAGAAATTTGTCATTGTCGCATCTCAGCAAAAACCTACACAGATTCAATCTTCGAGCGGTACGGGGCACATCGTAGTACACAGTTCGCAGCCAACCATAGTGAGGAGTCAAAACGTTCAGTCGGTGGTGGTGACGAGCGGACCGGCGCCGGGCGGCGCAGCGCAGAAGTTGATGGTCGTCGGAGTGCAGCCGCAGCACAACCAAGGACAGGTCAGCCAGGCGCCAGTGTCGGTGGTGGCGAAGCCCGTGTTCGCTCGTGGGGGCGGCGGACCACAACCTCCGCCTGAACTAGACGACCTCTCTCATCTCGCCTGA
Protein
MGDSELAYGSSLTLDSMKVIAESVGIATLGDDAAKELADDVTYRLKVIVQDAMKFMHHSKRQKLSITDIDHALKIKNNECQYGFIQPDSLPFRFASGGGRELHFIEEKEIDLSEILSAPPPKVPLDVSVRAHWLSVDGVQPTVPENPPPLSKEAQKLESVDPISKLSKPSNKDSAGKPICGKAARLKASESVHVKQLATHELSVEQQLYYKEITEACVGSDETRRAEALQSLACDPGLHEMLPRMCTFISEGVKVNVVQNNLALLIYLMRMVKALLDNQSLYLEKYLHELIPSVSTCIVSRQLCLRPEMDNHWALRDFAARLMAQICKTFNTSTNNLQTRVTRLFAKALQCPSQTNNETGPSMASMKESEKTPLASLYGAVQGLAELGPEVVKVFILPRVRWLGERVEGALSGIGGADRLAAGNLKHQLLKVLAPVVRQLRQPPDQPEDYKRDFGYLGPSLQQAVSKIRSSPSCSTAGGAVAVVTCTPPLLPTAPSPSPSNISHTTKTETVTTRNLVIASSTPQSPAPSTPPPQKFVIVASQQKPTQIQSSSGTGHIVVHSSQPTIVRSQNVQSVVVTSGPAPGGAAQKLMVVGVQPQHNQGQVSQAPVSVVAKPVFARGGGGPQPPPELDDLSHLA
Summary
Description
TFIID is a multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors.
Subunit
Belongs to the TFIID complex which is composed of TATA binding protein (Tbp) and a number of TBP-associated factors (TAFs). E(y)1 and Taf6 exist as a heterotetramer. Interacts with Taf1.
Similarity
Belongs to the TAF6 family.
Keywords
3D-structure
Complete proteome
Direct protein sequencing
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Transcription initiation factor TFIID subunit 6
Uniprot
H9JDI2
A0A2A4JYL1
A0A3S2M480
A0A212EL00
A0A2H1VC13
A0A194R779
+ More
A0A194PJ26 A0A1Q3F3Y4 B0W679 A0A182GHB5 A0A182G6Q3 Q173U6 A0A067RLB9 A0A1L8DGA6 A0A2J7PVD4 A0A1S4FF60 A0A1B0DDH3 A0A336LQS8 A0A182RX57 A0A2M4ASL3 A0A2M4BG90 A0A2M4AU65 A0A1I8PIW2 A0A182FPS3 W5J5A2 A0A182S8Q5 A0A182NCY6 A0A1A9WSM1 A0A182LZB0 A0A182K7V1 A0A182TL05 A0A1A9XY63 A0A1A9UFW5 A0A182USX6 T1PAC5 A0A1B0BNE1 A0A1I8N245 A0A1B0FPY1 A0A1A9ZKL8 A0A182QAX3 A0A182XG30 A0A182IW00 A0A182PS80 Q7QB06 A0A182L717 A0A182ID76 B3M6Z6 A0A0P8ZW36 A0A182W190 A0A1W4URL5 A0A1W4UTC9 A0A0M4EFL9 B4PGE8 A0A0R1E2E1 B4LCM3 B4KZC1 A0A0R1E3M3 E0VQC8 A0A0R1E3Y0 B3NIF8 A0A0Q9XDG4 A0A0Q5U5E2 A0A1B6INQ8 A0A0Q5UK57 A0A0V0G8C4 A0A0Q5U6E3 A0A0A1X2I7 A0A1B6F6H2 A0A1B6J6G3 M9PG32 P49847 W8BYF7 W8C4U0 A0A0P4VIX9 A0A1B6FS71 A0A069DZ89 A0A182Y5C2 A0A023FAE5 A0A0T6AYS9 A0A1B6MJX6 A0A1S4GKG7 A0A034W993 B4IY22 B4IIM9 B4QQV6 A0A0J9UPB2 A0A0K8U6I1 R4WL71 A0A1W4W5W9 A0A2P8ZHJ9 U4U1R4 N6THH7 D6WJN6 A0A146M2U4 U4U3V0 A0A1Y1LKX9 B4MXM4 T1HR29 A0A087ZWF1
A0A194PJ26 A0A1Q3F3Y4 B0W679 A0A182GHB5 A0A182G6Q3 Q173U6 A0A067RLB9 A0A1L8DGA6 A0A2J7PVD4 A0A1S4FF60 A0A1B0DDH3 A0A336LQS8 A0A182RX57 A0A2M4ASL3 A0A2M4BG90 A0A2M4AU65 A0A1I8PIW2 A0A182FPS3 W5J5A2 A0A182S8Q5 A0A182NCY6 A0A1A9WSM1 A0A182LZB0 A0A182K7V1 A0A182TL05 A0A1A9XY63 A0A1A9UFW5 A0A182USX6 T1PAC5 A0A1B0BNE1 A0A1I8N245 A0A1B0FPY1 A0A1A9ZKL8 A0A182QAX3 A0A182XG30 A0A182IW00 A0A182PS80 Q7QB06 A0A182L717 A0A182ID76 B3M6Z6 A0A0P8ZW36 A0A182W190 A0A1W4URL5 A0A1W4UTC9 A0A0M4EFL9 B4PGE8 A0A0R1E2E1 B4LCM3 B4KZC1 A0A0R1E3M3 E0VQC8 A0A0R1E3Y0 B3NIF8 A0A0Q9XDG4 A0A0Q5U5E2 A0A1B6INQ8 A0A0Q5UK57 A0A0V0G8C4 A0A0Q5U6E3 A0A0A1X2I7 A0A1B6F6H2 A0A1B6J6G3 M9PG32 P49847 W8BYF7 W8C4U0 A0A0P4VIX9 A0A1B6FS71 A0A069DZ89 A0A182Y5C2 A0A023FAE5 A0A0T6AYS9 A0A1B6MJX6 A0A1S4GKG7 A0A034W993 B4IY22 B4IIM9 B4QQV6 A0A0J9UPB2 A0A0K8U6I1 R4WL71 A0A1W4W5W9 A0A2P8ZHJ9 U4U1R4 N6THH7 D6WJN6 A0A146M2U4 U4U3V0 A0A1Y1LKX9 B4MXM4 T1HR29 A0A087ZWF1
Pubmed
19121390
22118469
26354079
26483478
17510324
24845553
+ More
20920257 23761445 25315136 12364791 20966253 17994087 17550304 20566863 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 7545910 8262073 18327897 8598927 24495485 27129103 26334808 25244985 25474469 25348373 22936249 23691247 29403074 23537049 18362917 19820115 26823975 28004739
20920257 23761445 25315136 12364791 20966253 17994087 17550304 20566863 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 7545910 8262073 18327897 8598927 24495485 27129103 26334808 25244985 25474469 25348373 22936249 23691247 29403074 23537049 18362917 19820115 26823975 28004739
EMBL
BABH01030138
BABH01030139
NWSH01000353
PCG77111.1
RSAL01000040
RVE50989.1
+ More
AGBW02014166 OWR42164.1 ODYU01001734 SOQ38341.1 KQ460615 KPJ13678.1 KQ459602 KPI93426.1 GFDL01012798 JAV22247.1 DS231847 EDS36426.1 JXUM01063366 KQ562245 KXJ76316.1 JXUM01045411 JXUM01045412 JXUM01045413 JXUM01045414 KQ561437 KXJ78595.1 CH477416 EAT41350.1 KK852443 KDR23828.1 GFDF01008623 JAV05461.1 NEVH01020964 PNF20294.1 AJVK01014493 UFQT01000118 SSX20406.1 GGFK01010401 MBW43722.1 GGFJ01002896 MBW52037.1 GGFK01011018 MBW44339.1 ADMH02002178 ETN58045.1 AXCM01000508 KA645644 AFP60273.1 JXJN01017347 CCAG010001380 AXCN02001560 AAAB01008880 EAA08678.5 APCN01005045 CH902618 EDV40861.1 KPU78819.1 CP012525 ALC44771.1 CM000159 EDW95307.1 KRK02371.1 CH940647 EDW69886.1 CH933809 EDW18947.1 KRK02369.1 DS235418 EEB15584.1 KRK02370.1 CH954178 EDV52383.1 KRG06429.1 KQS44248.1 GECU01019157 JAS88549.1 KQS44250.1 GECL01001851 JAP04273.1 KQS44249.1 GBXI01009322 JAD04970.1 GECZ01023951 JAS45818.1 GECU01012950 JAS94756.1 AE014296 AGB94749.1 AY051702 U06459 L25443 GAMC01002178 JAC04378.1 GAMC01002177 JAC04379.1 GDKW01002572 JAI54023.1 GECZ01016731 JAS53038.1 GBGD01001060 JAC87829.1 GBBI01000261 JAC18451.1 LJIG01022491 KRT80297.1 GEBQ01003818 JAT36159.1 GAKP01006811 JAC52141.1 CH916366 EDV97565.1 CH480844 EDW49800.1 CM000363 CM002912 EDX11094.1 KMZ00570.1 KMZ00571.1 GDHF01030128 JAI22186.1 AK418424 BAN21595.1 PYGN01000056 PSN55976.1 KB631899 ERL87002.1 APGK01037114 APGK01037115 KB740948 ENN77233.1 KQ971343 EFA03123.1 GDHC01005542 JAQ13087.1 ERL87003.1 GEZM01057579 JAV72246.1 CH963876 EDW76793.1 ACPB03009189
AGBW02014166 OWR42164.1 ODYU01001734 SOQ38341.1 KQ460615 KPJ13678.1 KQ459602 KPI93426.1 GFDL01012798 JAV22247.1 DS231847 EDS36426.1 JXUM01063366 KQ562245 KXJ76316.1 JXUM01045411 JXUM01045412 JXUM01045413 JXUM01045414 KQ561437 KXJ78595.1 CH477416 EAT41350.1 KK852443 KDR23828.1 GFDF01008623 JAV05461.1 NEVH01020964 PNF20294.1 AJVK01014493 UFQT01000118 SSX20406.1 GGFK01010401 MBW43722.1 GGFJ01002896 MBW52037.1 GGFK01011018 MBW44339.1 ADMH02002178 ETN58045.1 AXCM01000508 KA645644 AFP60273.1 JXJN01017347 CCAG010001380 AXCN02001560 AAAB01008880 EAA08678.5 APCN01005045 CH902618 EDV40861.1 KPU78819.1 CP012525 ALC44771.1 CM000159 EDW95307.1 KRK02371.1 CH940647 EDW69886.1 CH933809 EDW18947.1 KRK02369.1 DS235418 EEB15584.1 KRK02370.1 CH954178 EDV52383.1 KRG06429.1 KQS44248.1 GECU01019157 JAS88549.1 KQS44250.1 GECL01001851 JAP04273.1 KQS44249.1 GBXI01009322 JAD04970.1 GECZ01023951 JAS45818.1 GECU01012950 JAS94756.1 AE014296 AGB94749.1 AY051702 U06459 L25443 GAMC01002178 JAC04378.1 GAMC01002177 JAC04379.1 GDKW01002572 JAI54023.1 GECZ01016731 JAS53038.1 GBGD01001060 JAC87829.1 GBBI01000261 JAC18451.1 LJIG01022491 KRT80297.1 GEBQ01003818 JAT36159.1 GAKP01006811 JAC52141.1 CH916366 EDV97565.1 CH480844 EDW49800.1 CM000363 CM002912 EDX11094.1 KMZ00570.1 KMZ00571.1 GDHF01030128 JAI22186.1 AK418424 BAN21595.1 PYGN01000056 PSN55976.1 KB631899 ERL87002.1 APGK01037114 APGK01037115 KB740948 ENN77233.1 KQ971343 EFA03123.1 GDHC01005542 JAQ13087.1 ERL87003.1 GEZM01057579 JAV72246.1 CH963876 EDW76793.1 ACPB03009189
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000002320 UP000069940 UP000249989 UP000008820 UP000027135 UP000235965 UP000092462 UP000075900 UP000095300 UP000069272 UP000000673 UP000075901 UP000075884 UP000091820 UP000075883 UP000075881 UP000075902 UP000092443 UP000078200 UP000075903 UP000092460 UP000095301 UP000092444 UP000092445 UP000075886 UP000076407 UP000075880 UP000075885 UP000007062 UP000075882 UP000075840 UP000007801 UP000075920 UP000192221 UP000092553 UP000002282 UP000008792 UP000009192 UP000009046 UP000008711 UP000000803 UP000076408 UP000001070 UP000001292 UP000000304 UP000192223 UP000245037 UP000030742 UP000019118 UP000007266 UP000007798 UP000015103 UP000005203
UP000002320 UP000069940 UP000249989 UP000008820 UP000027135 UP000235965 UP000092462 UP000075900 UP000095300 UP000069272 UP000000673 UP000075901 UP000075884 UP000091820 UP000075883 UP000075881 UP000075902 UP000092443 UP000078200 UP000075903 UP000092460 UP000095301 UP000092444 UP000092445 UP000075886 UP000076407 UP000075880 UP000075885 UP000007062 UP000075882 UP000075840 UP000007801 UP000075920 UP000192221 UP000092553 UP000002282 UP000008792 UP000009192 UP000009046 UP000008711 UP000000803 UP000076408 UP000001070 UP000001292 UP000000304 UP000192223 UP000245037 UP000030742 UP000019118 UP000007266 UP000007798 UP000015103 UP000005203
Interpro
Gene 3D
ProteinModelPortal
H9JDI2
A0A2A4JYL1
A0A3S2M480
A0A212EL00
A0A2H1VC13
A0A194R779
+ More
A0A194PJ26 A0A1Q3F3Y4 B0W679 A0A182GHB5 A0A182G6Q3 Q173U6 A0A067RLB9 A0A1L8DGA6 A0A2J7PVD4 A0A1S4FF60 A0A1B0DDH3 A0A336LQS8 A0A182RX57 A0A2M4ASL3 A0A2M4BG90 A0A2M4AU65 A0A1I8PIW2 A0A182FPS3 W5J5A2 A0A182S8Q5 A0A182NCY6 A0A1A9WSM1 A0A182LZB0 A0A182K7V1 A0A182TL05 A0A1A9XY63 A0A1A9UFW5 A0A182USX6 T1PAC5 A0A1B0BNE1 A0A1I8N245 A0A1B0FPY1 A0A1A9ZKL8 A0A182QAX3 A0A182XG30 A0A182IW00 A0A182PS80 Q7QB06 A0A182L717 A0A182ID76 B3M6Z6 A0A0P8ZW36 A0A182W190 A0A1W4URL5 A0A1W4UTC9 A0A0M4EFL9 B4PGE8 A0A0R1E2E1 B4LCM3 B4KZC1 A0A0R1E3M3 E0VQC8 A0A0R1E3Y0 B3NIF8 A0A0Q9XDG4 A0A0Q5U5E2 A0A1B6INQ8 A0A0Q5UK57 A0A0V0G8C4 A0A0Q5U6E3 A0A0A1X2I7 A0A1B6F6H2 A0A1B6J6G3 M9PG32 P49847 W8BYF7 W8C4U0 A0A0P4VIX9 A0A1B6FS71 A0A069DZ89 A0A182Y5C2 A0A023FAE5 A0A0T6AYS9 A0A1B6MJX6 A0A1S4GKG7 A0A034W993 B4IY22 B4IIM9 B4QQV6 A0A0J9UPB2 A0A0K8U6I1 R4WL71 A0A1W4W5W9 A0A2P8ZHJ9 U4U1R4 N6THH7 D6WJN6 A0A146M2U4 U4U3V0 A0A1Y1LKX9 B4MXM4 T1HR29 A0A087ZWF1
A0A194PJ26 A0A1Q3F3Y4 B0W679 A0A182GHB5 A0A182G6Q3 Q173U6 A0A067RLB9 A0A1L8DGA6 A0A2J7PVD4 A0A1S4FF60 A0A1B0DDH3 A0A336LQS8 A0A182RX57 A0A2M4ASL3 A0A2M4BG90 A0A2M4AU65 A0A1I8PIW2 A0A182FPS3 W5J5A2 A0A182S8Q5 A0A182NCY6 A0A1A9WSM1 A0A182LZB0 A0A182K7V1 A0A182TL05 A0A1A9XY63 A0A1A9UFW5 A0A182USX6 T1PAC5 A0A1B0BNE1 A0A1I8N245 A0A1B0FPY1 A0A1A9ZKL8 A0A182QAX3 A0A182XG30 A0A182IW00 A0A182PS80 Q7QB06 A0A182L717 A0A182ID76 B3M6Z6 A0A0P8ZW36 A0A182W190 A0A1W4URL5 A0A1W4UTC9 A0A0M4EFL9 B4PGE8 A0A0R1E2E1 B4LCM3 B4KZC1 A0A0R1E3M3 E0VQC8 A0A0R1E3Y0 B3NIF8 A0A0Q9XDG4 A0A0Q5U5E2 A0A1B6INQ8 A0A0Q5UK57 A0A0V0G8C4 A0A0Q5U6E3 A0A0A1X2I7 A0A1B6F6H2 A0A1B6J6G3 M9PG32 P49847 W8BYF7 W8C4U0 A0A0P4VIX9 A0A1B6FS71 A0A069DZ89 A0A182Y5C2 A0A023FAE5 A0A0T6AYS9 A0A1B6MJX6 A0A1S4GKG7 A0A034W993 B4IY22 B4IIM9 B4QQV6 A0A0J9UPB2 A0A0K8U6I1 R4WL71 A0A1W4W5W9 A0A2P8ZHJ9 U4U1R4 N6THH7 D6WJN6 A0A146M2U4 U4U3V0 A0A1Y1LKX9 B4MXM4 T1HR29 A0A087ZWF1
PDB
6MZM
E-value=1.27131e-137,
Score=1257
Ontologies
GO
PANTHER
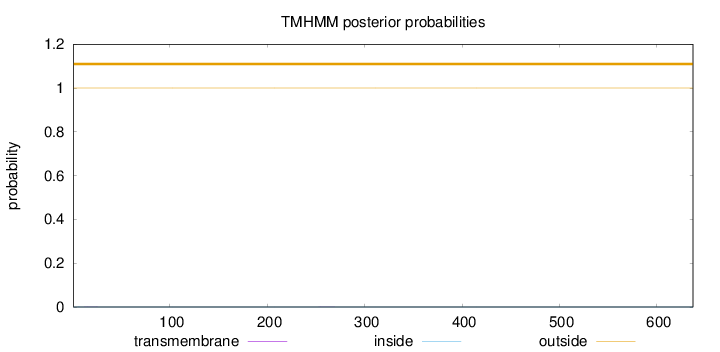
Topology
Subcellular location
Nucleus
Length:
637
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00686999999999999
Exp number, first 60 AAs:
0.00267
Total prob of N-in:
0.00036
outside
1 - 637
Population Genetic Test Statistics
Pi
257.17763
Theta
174.304337
Tajima's D
1.616782
CLR
0.358016
CSRT
0.808359582020899
Interpretation
Uncertain
