Pre Gene Modal
BGIBMGA007512
Annotation
PREDICTED:_UDP-glucose:glycoprotein_glucosyltransferase_[Bombyx_mori]
Full name
UDP-glucose:glycoprotein glucosyltransferase
Alternative Name
UDP--Glc:glycoprotein glucosyltransferase
Location in the cell
Cytoplasmic Reliability : 2.142
Sequence
CDS
ATGTATGGCGATAAAATAGCAGGAGTTTTAGACAAGAAAAAAGCGACCATTACTAACGAAGTTACCGATGATGACGATGAGGAGTCGGTAGAAGTGACGAGTGACATGCGTCTGAAGCTGGTGTCGGTGCTGGGAGGTCGCGAGTCCCGCGCCCGCACCGCGCTGCCGAGCGGACTACACACCGAACATTCGCTCATCGAACTGCCTCCTGTGTACGACAATGAAGCCGCCGTCGAGGTTGTCGCAGTGCTGGACCCGGCGTCGGGCGCGGCGCAACGCCTCGCGCCGCTGCTGCTCGTGCTACGTCGAGTCGTCAATTGTAATATTAAATTGTTCCTCAACCCCCAAGACAAGAACTCTGATATGCCACTGAAAAGCTTCTACCGGTACGTGTTGGAGCCGGAGCTGCAGTTCACGGCGGCGGGCGCGCTGTCGGGCGGCGCGCTGGCACGGTTCTCGCGGCTGCCGCACGCGCCGCTGCTGTCCATGGAGCTGCGCACGCCGACCAACTGGCTCGTGGAGTGCGTCAAGTCCGTCTACGACCTCGACAACATCAGGCTCGCCGATGTCGAAACCGTCGTGCACAGCGAATTCGAACTAGAATACCTGCTGTTAGAAGGTCACGCTTGGGACACAACTCTCGGTACGCCACCTCGAGGTCTACAGCTGATACTTGGGACCAGGGAGAAACCCGAACTCATGGACACCATTGTGATGGCGAACCTGGGCTACTTCCAGCTGAAAGCCAACCCAGGGGCGTGGACGTTGCGCCTGCGACCCGGGCGCTCTGAAGAAATTTATGAGATTGTTGGGCACGAAAACACGGACACGCCGGCGGGCAGCCATGACATACAGGTGCTGATGAGCTCGTTCCGCAGCCACGTCATCAAGTTGAGGGTGAGCAAGAAGCCAGACAAGCAACACCTGGATTTGCTCGTCGAAAATGACGACAAAAACTCAGGAGGACTCTGGAATTCCATTGCGAGCTCGTTCGGTGGTGGAGACGAATCTGAAGCTCAAGACGACACGATCAACGTGTTCTCAGTGGCTTCAGGACATTTGTATGAAAGATTCCTTCGGATTATGATGTTGTCTGTTCTTAAAAACACAAAATCACCCGTCAAGTTCTGGTTTCTCAAGAATTACCTCAGTCCATCGCTTAAGGATATTCTACCATACATGGCCCAAGAATATGGGTTCGAGTACGAGCTGGTCCAGTACCAGTGGCCGCGCTGGCTGCAGCGACAACGCGACCGGCAACGCACCATCTGGGGATACAAGATCCTGTTCCTGGACGTGTTGTTCCCGCTGCACGTCAAGAAGATTATCTTCGTCGATGCCGACCAGATTGTTCGTGCTGATCTCAAAGAATTGGTGGAATTAGATTTAGGAGGCGCACCATACGGTTATACACCGTTCTGTGATAGTAGGACAGAAATGGATGGTTTCAGGTTCTGGAAGCAAGGCTACTGGCGCAATCACCTTCAAGGTCGTAGCTACCACATCAGCGCGCTGTACGTCGTGGACTTGAAACGGTTCCGCCGCATCGCCGCGGGGGACCGCCTCCGAGGACAGTACCAGGCGCTCAGTCAAGACCCTAACAGCTTGTCTAATTTAGATCAAGATTTACCCAACAACATGATCCATCAAGTGGCCATCAAATCGTTACCGCAAGAGTGGCTGTGGTGTGAGACCTGGTGTGACGACGACTCGAAGAAATACGCCAAAACAATTGACTTGTGCAACAACCCTATGACTAAAGAAGCCAAGCTCTCCGCTGCGATGCGCATAGTACCAGAATGGAAAGAATACGACGGTGAAGTGAAGGCTTTACAGGCGCGTGTTAGACGAGGTCTCTACCAATCTGCTGACCATGAACAGGTATAA
Protein
MYGDKIAGVLDKKKATITNEVTDDDDEESVEVTSDMRLKLVSVLGGRESRARTALPSGLHTEHSLIELPPVYDNEAAVEVVAVLDPASGAAQRLAPLLLVLRRVVNCNIKLFLNPQDKNSDMPLKSFYRYVLEPELQFTAAGALSGGALARFSRLPHAPLLSMELRTPTNWLVECVKSVYDLDNIRLADVETVVHSEFELEYLLLEGHAWDTTLGTPPRGLQLILGTREKPELMDTIVMANLGYFQLKANPGAWTLRLRPGRSEEIYEIVGHENTDTPAGSHDIQVLMSSFRSHVIKLRVSKKPDKQHLDLLVENDDKNSGGLWNSIASSFGGGDESEAQDDTINVFSVASGHLYERFLRIMMLSVLKNTKSPVKFWFLKNYLSPSLKDILPYMAQEYGFEYELVQYQWPRWLQRQRDRQRTIWGYKILFLDVLFPLHVKKIIFVDADQIVRADLKELVELDLGGAPYGYTPFCDSRTEMDGFRFWKQGYWRNHLQGRSYHISALYVVDLKRFRRIAAGDRLRGQYQALSQDPNSLSNLDQDLPNNMIHQVAIKSLPQEWLWCETWCDDDSKKYAKTIDLCNNPMTKEAKLSAAMRIVPEWKEYDGEVKALQARVRRGLYQSADHEQV
Summary
Subunit
Monomer.
Similarity
Belongs to the glycosyltransferase 8 family.
Keywords
Complete proteome
Direct protein sequencing
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Reference proteome
Signal
Transferase
Feature
chain UDP-glucose:glycoprotein glucosyltransferase
Uniprot
H9JDB5
A0A3S2PGS0
A0A2A4JZ71
A0A212EKZ9
S4NNL1
A0A194PK85
+ More
A0A2H1WXD8 A0A1W4XG73 A0A1B6F0P2 A0A1W4X5K4 A0A2P8YV46 A0A2J7QLD5 A0A1B6BXN7 D6WKY8 A0A139WGR5 A0A0A9Y4P1 A0A0K8S9E8 A0A023F3E2 E2AEM6 A0A026X1D2 A0A3L8DM42 A0A224XGM8 Q09332 B3M861 A0A2K8JM31 B4N4V0 E9JAG6 A0A0J9RXZ5 B4QPX4 A0A1W4UQX1 A0A151WG09 B3NDU6 A0A151I9B4 A0A0J7L7L7 A0A3B0JS60 A0A1Y1LFV4 A0A151J135 A0A232F7H1 B4LD55 B4PH35 A0A195FXM8 E2C511 Q2M0E2 A0A1B0G8K0 K7IP78 A0A1A9V3T6 A0A0M8ZQZ6 A0A1B0AI77 B4KZ01 A0A195BU08 A0A0L0C6E8 B4GRR3 J9JSC5 A0A0P5S3R7 A0A2S2PJX3 F4X2J1 A0A0L7RIX3 A0A158NV88 A0A0P5SJD5 A0A2S2RAE3 A0A1A9W760 W4VRL2 B4IYE1 A0A1A9Y0W5 A0A1B0B6V6 Q17PC9 U4U2M4 A0A0C9R065 N6UD09 A0A087US98 A0A0M5IYM8 E9GSF3 A0A1I8PAU5 A0A0P5U1G1 A0A0P6G1Y1 A0A0P6HHC1 A0A0P5SER3 A0A0N8EEV8 A0A0P5YC41 A0A0N8BTT2 A0A0P5FUJ3 A0A0P5S5H5 A0A0P5IHT0 A0A0P5WAX1 A0A0P4XYG5 A0A0P5UC95 A0A0N8BS75 A0A0P5SM18 A0A1J1IZW3 A0A0P5MRT7 T1HRF6 A0A0P5ZJ44 A0A0P6EZK3 B0WIX0 A0A182RZF1 A0A0P5TVR6 A0A182VQ37 A0A088AHX7 A0A1Q3FBA7 A0A1Q3FBB8
A0A2H1WXD8 A0A1W4XG73 A0A1B6F0P2 A0A1W4X5K4 A0A2P8YV46 A0A2J7QLD5 A0A1B6BXN7 D6WKY8 A0A139WGR5 A0A0A9Y4P1 A0A0K8S9E8 A0A023F3E2 E2AEM6 A0A026X1D2 A0A3L8DM42 A0A224XGM8 Q09332 B3M861 A0A2K8JM31 B4N4V0 E9JAG6 A0A0J9RXZ5 B4QPX4 A0A1W4UQX1 A0A151WG09 B3NDU6 A0A151I9B4 A0A0J7L7L7 A0A3B0JS60 A0A1Y1LFV4 A0A151J135 A0A232F7H1 B4LD55 B4PH35 A0A195FXM8 E2C511 Q2M0E2 A0A1B0G8K0 K7IP78 A0A1A9V3T6 A0A0M8ZQZ6 A0A1B0AI77 B4KZ01 A0A195BU08 A0A0L0C6E8 B4GRR3 J9JSC5 A0A0P5S3R7 A0A2S2PJX3 F4X2J1 A0A0L7RIX3 A0A158NV88 A0A0P5SJD5 A0A2S2RAE3 A0A1A9W760 W4VRL2 B4IYE1 A0A1A9Y0W5 A0A1B0B6V6 Q17PC9 U4U2M4 A0A0C9R065 N6UD09 A0A087US98 A0A0M5IYM8 E9GSF3 A0A1I8PAU5 A0A0P5U1G1 A0A0P6G1Y1 A0A0P6HHC1 A0A0P5SER3 A0A0N8EEV8 A0A0P5YC41 A0A0N8BTT2 A0A0P5FUJ3 A0A0P5S5H5 A0A0P5IHT0 A0A0P5WAX1 A0A0P4XYG5 A0A0P5UC95 A0A0N8BS75 A0A0P5SM18 A0A1J1IZW3 A0A0P5MRT7 T1HRF6 A0A0P5ZJ44 A0A0P6EZK3 B0WIX0 A0A182RZF1 A0A0P5TVR6 A0A182VQ37 A0A088AHX7 A0A1Q3FBA7 A0A1Q3FBB8
EC Number
2.4.1.-
Pubmed
EMBL
BABH01030146
RSAL01000040
RVE50987.1
NWSH01000353
PCG77109.1
AGBW02014166
+ More
OWR42166.1 GAIX01012219 JAA80341.1 KQ459602 KPI93423.1 ODYU01011743 SOQ57662.1 GECZ01025957 GECZ01006960 JAS43812.1 JAS62809.1 PYGN01000342 PSN48119.1 NEVH01013247 PNF29392.1 GEDC01031519 GEDC01023173 JAS05779.1 JAS14125.1 KQ971343 EFA03542.1 KYB27153.1 GBHO01017517 GBHO01017516 GDHC01020361 JAG26087.1 JAG26088.1 JAP98267.1 GBRD01016451 JAG49375.1 GBBI01002951 JAC15761.1 GL438870 EFN68101.1 KK107039 EZA61903.1 QOIP01000006 RLU21252.1 GFTR01008816 JAW07610.1 U20554 AE014296 AY058353 BT044158 CH902618 EDV41000.1 KY031137 ATU82888.1 CH964101 EDW79389.1 GL770322 EFZ10193.1 CM002912 KMZ00458.1 CM000363 EDX11002.1 KQ983203 KYQ46715.1 CH954178 EDV52300.1 KQ978330 KYM95086.1 LBMM01000270 KMR02238.1 OUUW01000002 SPP76196.1 GEZM01060864 JAV70845.1 KQ980575 KYN15496.1 NNAY01000809 OXU26398.1 CH940647 EDW69936.1 CM000159 EDW95405.1 KQ981193 KYN45181.1 GL452714 EFN76965.1 CH379069 EAL30992.2 CCAG010000259 KQ435969 KOX67853.1 CH933809 EDW18893.1 KQ976417 KYM89904.1 JRES01000836 KNC27850.1 CH479188 EDW40448.1 ABLF02034488 ABLF02034494 GDIQ01092774 JAL58952.1 GGMR01017086 MBY29705.1 GL888584 EGI59316.1 KQ414583 KOC70784.1 ADTU01027005 ADTU01027006 GDIQ01094920 JAL56806.1 GGMS01017735 MBY86938.1 GANO01002072 JAB57799.1 CH916366 EDV97614.1 JXJN01009254 CH477193 EAT48502.1 KB631930 ERL87312.1 GBYB01009499 JAG79266.1 APGK01028346 APGK01028347 APGK01028348 KB740686 ENN79555.1 KK121329 KFM80237.1 CP012525 ALC43831.1 GL732562 EFX77585.1 GDIP01121068 JAL82646.1 GDIQ01039744 JAN54993.1 GDIQ01028200 JAN66537.1 GDIQ01092775 JAL58951.1 GDIQ01033604 JAN61133.1 GDIP01060578 GDIP01045447 LRGB01001663 JAM43137.1 KZS10926.1 GDIQ01245024 GDIQ01239529 GDIQ01145603 JAL06123.1 GDIQ01253528 GDIQ01253527 GDIQ01253526 GDIQ01252373 GDIQ01252372 GDIQ01252371 GDIQ01250875 GDIQ01250874 GDIQ01249625 GDIQ01249624 GDIQ01247928 GDIQ01247927 GDIQ01204635 GDIQ01204634 GDIQ01202836 GDIQ01202835 JAJ98197.1 GDIQ01239530 GDIQ01239528 GDIQ01208783 GDIQ01208782 GDIQ01208781 GDIQ01207536 GDIQ01207535 GDIQ01207534 GDIQ01206134 GDIQ01206133 GDIQ01185837 GDIQ01159698 GDIQ01157854 GDIQ01132518 GDIQ01132517 GDIQ01125561 GDIQ01110618 GDIQ01101029 GDIQ01101028 GDIQ01083179 GDIQ01068489 GDIQ01065640 GDIQ01065639 GDIQ01033603 JAL50698.1 GDIQ01214127 JAK37598.1 GDIP01089428 JAM14287.1 GDIP01235108 JAI88293.1 GDIP01115123 JAL88591.1 GDIQ01150059 JAL01667.1 GDIP01138077 JAL65637.1 CVRI01000065 CRL05679.1 GDIQ01152905 JAK98820.1 ACPB03009186 GDIP01042829 JAM60886.1 GDIQ01055795 JAN38942.1 DS231953 EDS28796.1 GDIP01126026 JAL77688.1 GFDL01010181 JAV24864.1 GFDL01010195 JAV24850.1
OWR42166.1 GAIX01012219 JAA80341.1 KQ459602 KPI93423.1 ODYU01011743 SOQ57662.1 GECZ01025957 GECZ01006960 JAS43812.1 JAS62809.1 PYGN01000342 PSN48119.1 NEVH01013247 PNF29392.1 GEDC01031519 GEDC01023173 JAS05779.1 JAS14125.1 KQ971343 EFA03542.1 KYB27153.1 GBHO01017517 GBHO01017516 GDHC01020361 JAG26087.1 JAG26088.1 JAP98267.1 GBRD01016451 JAG49375.1 GBBI01002951 JAC15761.1 GL438870 EFN68101.1 KK107039 EZA61903.1 QOIP01000006 RLU21252.1 GFTR01008816 JAW07610.1 U20554 AE014296 AY058353 BT044158 CH902618 EDV41000.1 KY031137 ATU82888.1 CH964101 EDW79389.1 GL770322 EFZ10193.1 CM002912 KMZ00458.1 CM000363 EDX11002.1 KQ983203 KYQ46715.1 CH954178 EDV52300.1 KQ978330 KYM95086.1 LBMM01000270 KMR02238.1 OUUW01000002 SPP76196.1 GEZM01060864 JAV70845.1 KQ980575 KYN15496.1 NNAY01000809 OXU26398.1 CH940647 EDW69936.1 CM000159 EDW95405.1 KQ981193 KYN45181.1 GL452714 EFN76965.1 CH379069 EAL30992.2 CCAG010000259 KQ435969 KOX67853.1 CH933809 EDW18893.1 KQ976417 KYM89904.1 JRES01000836 KNC27850.1 CH479188 EDW40448.1 ABLF02034488 ABLF02034494 GDIQ01092774 JAL58952.1 GGMR01017086 MBY29705.1 GL888584 EGI59316.1 KQ414583 KOC70784.1 ADTU01027005 ADTU01027006 GDIQ01094920 JAL56806.1 GGMS01017735 MBY86938.1 GANO01002072 JAB57799.1 CH916366 EDV97614.1 JXJN01009254 CH477193 EAT48502.1 KB631930 ERL87312.1 GBYB01009499 JAG79266.1 APGK01028346 APGK01028347 APGK01028348 KB740686 ENN79555.1 KK121329 KFM80237.1 CP012525 ALC43831.1 GL732562 EFX77585.1 GDIP01121068 JAL82646.1 GDIQ01039744 JAN54993.1 GDIQ01028200 JAN66537.1 GDIQ01092775 JAL58951.1 GDIQ01033604 JAN61133.1 GDIP01060578 GDIP01045447 LRGB01001663 JAM43137.1 KZS10926.1 GDIQ01245024 GDIQ01239529 GDIQ01145603 JAL06123.1 GDIQ01253528 GDIQ01253527 GDIQ01253526 GDIQ01252373 GDIQ01252372 GDIQ01252371 GDIQ01250875 GDIQ01250874 GDIQ01249625 GDIQ01249624 GDIQ01247928 GDIQ01247927 GDIQ01204635 GDIQ01204634 GDIQ01202836 GDIQ01202835 JAJ98197.1 GDIQ01239530 GDIQ01239528 GDIQ01208783 GDIQ01208782 GDIQ01208781 GDIQ01207536 GDIQ01207535 GDIQ01207534 GDIQ01206134 GDIQ01206133 GDIQ01185837 GDIQ01159698 GDIQ01157854 GDIQ01132518 GDIQ01132517 GDIQ01125561 GDIQ01110618 GDIQ01101029 GDIQ01101028 GDIQ01083179 GDIQ01068489 GDIQ01065640 GDIQ01065639 GDIQ01033603 JAL50698.1 GDIQ01214127 JAK37598.1 GDIP01089428 JAM14287.1 GDIP01235108 JAI88293.1 GDIP01115123 JAL88591.1 GDIQ01150059 JAL01667.1 GDIP01138077 JAL65637.1 CVRI01000065 CRL05679.1 GDIQ01152905 JAK98820.1 ACPB03009186 GDIP01042829 JAM60886.1 GDIQ01055795 JAN38942.1 DS231953 EDS28796.1 GDIP01126026 JAL77688.1 GFDL01010181 JAV24864.1 GFDL01010195 JAV24850.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000192223
+ More
UP000245037 UP000235965 UP000007266 UP000000311 UP000053097 UP000279307 UP000000803 UP000007801 UP000007798 UP000000304 UP000192221 UP000075809 UP000008711 UP000078542 UP000036403 UP000268350 UP000078492 UP000215335 UP000008792 UP000002282 UP000078541 UP000008237 UP000001819 UP000092444 UP000002358 UP000078200 UP000053105 UP000092445 UP000009192 UP000078540 UP000037069 UP000008744 UP000007819 UP000007755 UP000053825 UP000005205 UP000091820 UP000001070 UP000092443 UP000092460 UP000008820 UP000030742 UP000019118 UP000054359 UP000092553 UP000000305 UP000095300 UP000076858 UP000183832 UP000015103 UP000002320 UP000075900 UP000075920 UP000005203
UP000245037 UP000235965 UP000007266 UP000000311 UP000053097 UP000279307 UP000000803 UP000007801 UP000007798 UP000000304 UP000192221 UP000075809 UP000008711 UP000078542 UP000036403 UP000268350 UP000078492 UP000215335 UP000008792 UP000002282 UP000078541 UP000008237 UP000001819 UP000092444 UP000002358 UP000078200 UP000053105 UP000092445 UP000009192 UP000078540 UP000037069 UP000008744 UP000007819 UP000007755 UP000053825 UP000005205 UP000091820 UP000001070 UP000092443 UP000092460 UP000008820 UP000030742 UP000019118 UP000054359 UP000092553 UP000000305 UP000095300 UP000076858 UP000183832 UP000015103 UP000002320 UP000075900 UP000075920 UP000005203
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JDB5
A0A3S2PGS0
A0A2A4JZ71
A0A212EKZ9
S4NNL1
A0A194PK85
+ More
A0A2H1WXD8 A0A1W4XG73 A0A1B6F0P2 A0A1W4X5K4 A0A2P8YV46 A0A2J7QLD5 A0A1B6BXN7 D6WKY8 A0A139WGR5 A0A0A9Y4P1 A0A0K8S9E8 A0A023F3E2 E2AEM6 A0A026X1D2 A0A3L8DM42 A0A224XGM8 Q09332 B3M861 A0A2K8JM31 B4N4V0 E9JAG6 A0A0J9RXZ5 B4QPX4 A0A1W4UQX1 A0A151WG09 B3NDU6 A0A151I9B4 A0A0J7L7L7 A0A3B0JS60 A0A1Y1LFV4 A0A151J135 A0A232F7H1 B4LD55 B4PH35 A0A195FXM8 E2C511 Q2M0E2 A0A1B0G8K0 K7IP78 A0A1A9V3T6 A0A0M8ZQZ6 A0A1B0AI77 B4KZ01 A0A195BU08 A0A0L0C6E8 B4GRR3 J9JSC5 A0A0P5S3R7 A0A2S2PJX3 F4X2J1 A0A0L7RIX3 A0A158NV88 A0A0P5SJD5 A0A2S2RAE3 A0A1A9W760 W4VRL2 B4IYE1 A0A1A9Y0W5 A0A1B0B6V6 Q17PC9 U4U2M4 A0A0C9R065 N6UD09 A0A087US98 A0A0M5IYM8 E9GSF3 A0A1I8PAU5 A0A0P5U1G1 A0A0P6G1Y1 A0A0P6HHC1 A0A0P5SER3 A0A0N8EEV8 A0A0P5YC41 A0A0N8BTT2 A0A0P5FUJ3 A0A0P5S5H5 A0A0P5IHT0 A0A0P5WAX1 A0A0P4XYG5 A0A0P5UC95 A0A0N8BS75 A0A0P5SM18 A0A1J1IZW3 A0A0P5MRT7 T1HRF6 A0A0P5ZJ44 A0A0P6EZK3 B0WIX0 A0A182RZF1 A0A0P5TVR6 A0A182VQ37 A0A088AHX7 A0A1Q3FBA7 A0A1Q3FBB8
A0A2H1WXD8 A0A1W4XG73 A0A1B6F0P2 A0A1W4X5K4 A0A2P8YV46 A0A2J7QLD5 A0A1B6BXN7 D6WKY8 A0A139WGR5 A0A0A9Y4P1 A0A0K8S9E8 A0A023F3E2 E2AEM6 A0A026X1D2 A0A3L8DM42 A0A224XGM8 Q09332 B3M861 A0A2K8JM31 B4N4V0 E9JAG6 A0A0J9RXZ5 B4QPX4 A0A1W4UQX1 A0A151WG09 B3NDU6 A0A151I9B4 A0A0J7L7L7 A0A3B0JS60 A0A1Y1LFV4 A0A151J135 A0A232F7H1 B4LD55 B4PH35 A0A195FXM8 E2C511 Q2M0E2 A0A1B0G8K0 K7IP78 A0A1A9V3T6 A0A0M8ZQZ6 A0A1B0AI77 B4KZ01 A0A195BU08 A0A0L0C6E8 B4GRR3 J9JSC5 A0A0P5S3R7 A0A2S2PJX3 F4X2J1 A0A0L7RIX3 A0A158NV88 A0A0P5SJD5 A0A2S2RAE3 A0A1A9W760 W4VRL2 B4IYE1 A0A1A9Y0W5 A0A1B0B6V6 Q17PC9 U4U2M4 A0A0C9R065 N6UD09 A0A087US98 A0A0M5IYM8 E9GSF3 A0A1I8PAU5 A0A0P5U1G1 A0A0P6G1Y1 A0A0P6HHC1 A0A0P5SER3 A0A0N8EEV8 A0A0P5YC41 A0A0N8BTT2 A0A0P5FUJ3 A0A0P5S5H5 A0A0P5IHT0 A0A0P5WAX1 A0A0P4XYG5 A0A0P5UC95 A0A0N8BS75 A0A0P5SM18 A0A1J1IZW3 A0A0P5MRT7 T1HRF6 A0A0P5ZJ44 A0A0P6EZK3 B0WIX0 A0A182RZF1 A0A0P5TVR6 A0A182VQ37 A0A088AHX7 A0A1Q3FBA7 A0A1Q3FBB8
PDB
5NV4
E-value=2.48216e-158,
Score=1435
Ontologies
GO
PANTHER
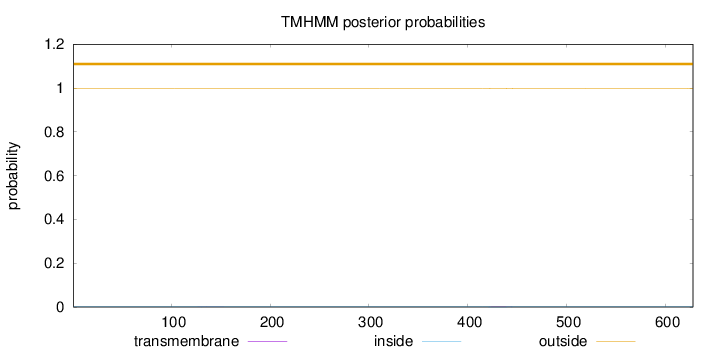
Topology
Subcellular location
Endoplasmic reticulum lumen
Length:
628
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03642
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00107
outside
1 - 628
Population Genetic Test Statistics
Pi
329.667883
Theta
176.708329
Tajima's D
2.433726
CLR
0.294697
CSRT
0.935053247337633
Interpretation
Uncertain
