Gene
KWMTBOMO09076 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007512
Annotation
PREDICTED:_UDP-glucose:glycoprotein_glucosyltransferase_[Bombyx_mori]
Full name
UDP-glucose:glycoprotein glucosyltransferase
Alternative Name
UDP--Glc:glycoprotein glucosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.473
Sequence
CDS
ATGATGAAAATCCCCGGCATAAGACTGTCTTTAGTGGTTTTAGCAATTGCTGTTGTGTCAGTGGCGTCAAATATAGATCCCATTAAACGAGATGAAAAGAAATCAAAATCTGTTACGACTTTTGTTAGTGCTAAATGGGAAGCTACACCTATTGTATTAGAACTGGCCGAGTATTTAGCTGGCGAAAGTTCAGATTTGTTTTGGTCTTTCGTTGACGGTATTAATTCTCTAAAATCTAGTCTTGACTCATTAGAAAATGACAAACAAATTTATGATGCCTGTATTGGAGTTGCTAGTACACTATTGGCTCCGGCACAACTTAGAATGGCTAAACTTGCACTTTCTATGCACACTACTTCACCAACAGTGCGTATGTTTGATCAAATAGCTATCCAGAACGGAGCTAAAGAAATTCAGTGCGACACATTTGTTAACATAGCAACAAACAATATTTGTGACAATGATGCGTTGAGAGATATACTTCAAACACATAATCTTAAAAATGTTGGTGATCAGAACAAAGAAATATATCTTCTGGACCATACTCATCCAAGTAGTGACAACAAAAGTATAACTGCCATTCTTTATGGGGAGCTTGGAACTAAGGATTTTGCAATGAAACACAAAATTTTGGCTGAATATGCTGACAAAAGTGCCATTAACTATGTTGTCAGATGGAACATCAAGACTCGTGGAAAACCTAAACTACGTCTATCAGGCTATGGAGTTGAACTTCAGTTAAAGAGTACAGAGTATAAAAGCCAAGACGATTCTACTCCAAAGGAAACAGAAGACAAAGAAACAGCAGCACAATCAGAGGAAGAGGATGAAAATGATCCCCAGAATCAGATTGATGGATTTAATTTTGGTAGATTAAAGAACCTATTTCCTGCTTTACGAACACCATTGGAACGGTTTCGACGGCACTTATCTGAATCTAGTGAAGAACTGGCCCCTCTGAAAGTATGGCAGATGCAGGCATTGAGTATGCAAGCTGCCGCTGCAGTTGTGGATGCGAATGATGCTGGTGGAGATGAAGCTTTGAGAGTTCTCACATCTATAGCTCAGAATTTTCCAATGCAGACAAAGTCTCTTATACACGTCAGTGTCCCGCGCGCTTTCCGCGATGAGGTACTGTACAACCAAGACATATGGGCATCAGCATTGGGGCTGCGGCCAGCTGAACCGTTGTTGTTAGTGTCAGGAGCTCAGTATGACGCTGAAGATGTTGATGTGATGGCTTTGCTTGCTGCATTGAGAGAAGACATTGGGCCAATGAATACATTACATGCTCTAGGTCTTCCTAAAAAGTTAATACATACTCTCATGTCATTGGATTTGGGCGAAAGTTACACCTGGGAAGAATATGGTCTTGATATCCGTGATACATCAATAACTTGGCTCAACGATCTGGAGTCTGACGATAGGTACCGCAGATGGCCTTCTTCGTTTATGGAATTGCTTCGGCCCACATATCCTGGCATGTTGAGAAATCTCAGACGAAACATATATAACTATGTCATCATAATAGACCCCACATCGGTACAATCTGGTCCACCGTTAAAGTTGGGTGAAACGTTGTTGAAGCATGCGACTCCAGTGCGCGTTGGGTTAGTCCTAGCGTCAACAAAAGGAGACGCGTCTTTAGAAGCAGCAGTCAGGTCCGCATTTAACTACATAGCACAGGAAAAGAATTCCAATAAAGAGGCATATTATTTCCTTACTCAGTTACTGAATCCTTCTGGTGATGACGTATTAGATATTAATTATGTGAAGAAACAATTGAAGCGACACGCGACTTCTGGTGCCAGTGTTGAAGACATTATTTCAGAAGATTCCGAATATAACTTTGGACGTCAAATCTCAGAAGAATTTGTATCTAAACTTGGCAGTGACAAGTATCCGCAGGTTTTGATAAATGGAGTACCACTAATGGACGAAGGCCCAACGCCGGTGACGTCGTCGGTCGAGTTACTCGAGGAGTCGCTGGTGACGTCACTGTCTCGCCACACAGCTCGCTTGCAGCGTGCTGTGTTCCGTGGTGAATTGTCCGATATGGATGACGCTGTTGATTACTTGATGAAACAGTCCCATATTATGCCACGGCTCAACCGTCGAATCCTCAGTTCAGAAACTTCGCAATATTTGGATTTGACCGGTGCCTCTTCACCTGCCGATTTGTTTACAGAAGATAAAATTCACAAACTATTACATTTAACAGGACGTGACGCGTTAGCGACTGCTCTACCAATTTTGAAGTACTTTTATAAACCCGGTAAAGTTGAAAAGAAGATAACGCAAACTATTTGGGTGATCGGAGATTTGAACAACCACGAATCCCGGGGACTATTGAGAAACGCTCTTACGTTTATGAGAGAATCCAGTGGCGTGAGAGTAGCGTTCATACCAAACGTGGACAGCTCAAACGATCAGTCTCTGAATAAAGTAGTGGTCGCGGCTCTTACGACGCTCGAGTCTACGGAAGCCACTAAATATGTTGTAAAATTATTAGAAGACGAAGGTTGTCATGAGAGGAAAGACTGTGATATTTTGAACGATATAAGGAATTCCTAA
Protein
MMKIPGIRLSLVVLAIAVVSVASNIDPIKRDEKKSKSVTTFVSAKWEATPIVLELAEYLAGESSDLFWSFVDGINSLKSSLDSLENDKQIYDACIGVASTLLAPAQLRMAKLALSMHTTSPTVRMFDQIAIQNGAKEIQCDTFVNIATNNICDNDALRDILQTHNLKNVGDQNKEIYLLDHTHPSSDNKSITAILYGELGTKDFAMKHKILAEYADKSAINYVVRWNIKTRGKPKLRLSGYGVELQLKSTEYKSQDDSTPKETEDKETAAQSEEEDENDPQNQIDGFNFGRLKNLFPALRTPLERFRRHLSESSEELAPLKVWQMQALSMQAAAAVVDANDAGGDEALRVLTSIAQNFPMQTKSLIHVSVPRAFRDEVLYNQDIWASALGLRPAEPLLLVSGAQYDAEDVDVMALLAALREDIGPMNTLHALGLPKKLIHTLMSLDLGESYTWEEYGLDIRDTSITWLNDLESDDRYRRWPSSFMELLRPTYPGMLRNLRRNIYNYVIIIDPTSVQSGPPLKLGETLLKHATPVRVGLVLASTKGDASLEAAVRSAFNYIAQEKNSNKEAYYFLTQLLNPSGDDVLDINYVKKQLKRHATSGASVEDIISEDSEYNFGRQISEEFVSKLGSDKYPQVLINGVPLMDEGPTPVTSSVELLEESLVTSLSRHTARLQRAVFRGELSDMDDAVDYLMKQSHIMPRLNRRILSSETSQYLDLTGASSPADLFTEDKIHKLLHLTGRDALATALPILKYFYKPGKVEKKITQTIWVIGDLNNHESRGLLRNALTFMRESSGVRVAFIPNVDSSNDQSLNKVVVAALTTLESTEATKYVVKLLEDEGCHERKDCDILNDIRNS
Summary
Subunit
Monomer.
Similarity
Belongs to the glycosyltransferase 8 family.
Keywords
Complete proteome
Direct protein sequencing
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Reference proteome
Signal
Transferase
Feature
chain UDP-glucose:glycoprotein glucosyltransferase
Uniprot
A0A2A4JZ71
A0A3S2PGS0
A0A194PK85
A0A212FIU0
A0A194R8U7
V5I9A4
+ More
A0A1W4XG73 A0A1W4X5K4 U4U2M4 D6WKY8 N6UD09 A0A139WGR5 A0A1B6JT93 E0VWP7 A0A182GVT0 A0A1Y1LFV4 A0A2J7QLD5 A0A023EXD3 A0A1Q3FBA7 A0A1Q3FBF1 A0A1Q3FBB8 B0WIX0 A0A2S2QB88 Q17PC9 A0A1A9W760 A0A2M3Z0E1 A0A182QRW2 A0A182NSP3 A0A182LTU5 A0A2H8TN89 A0A2M4B8X7 A0A2M4B9I4 A0A182TQR4 A0A232F7H1 A0A1B0B6V6 A0A1A9Y0W5 A0A182UZT9 A0A2P8YV46 Q7QAS6 A0A182KXF2 A0A182HZ93 A0A084VDY2 A0A1B0CKK3 A0A1B6BXN7 A0A2M3YZF2 A0A182Y7E9 A0A1B0AI77 A0A1A9V3T6 A0A182WVU5 W4VRL2 A0A1B0G8K0 K7IP78 T1PM16 A0A2M3ZZJ0 A0A182RZF1 A0A0L0C6E8 A0A2M3ZZJ9 A0A182F6N1 A0A182J6H8 A0A336KG93 W5JE34 A0A1I8PAU5 A0A034W5E6 E2AEM6 A0A1L8E268 A0A3L8DM42 A0A1L8E1R7 J9JSC5 A0A2K8JM31 A0A0K8V5M4 A0A182JZ54 A0A0M8ZQZ6 A0A0P4VK86 B4QPX4 A0A1B0DJK7 F4X2J1 A0A0J9RXZ5 A0A3B0JS60 A0A088AHX7 A0A158NV88 A0A151WG09 A0A182TAR5 B4IFW7 A0A182PH54 Q2M0E2 A0A195FXM8 B4N4V0 A0A0M5IYM8 A0A023F3E2 A0A0A1XG58 A0A224XGM8 B4IYE1 A0A1W4UQX1 W8AWU6 A0A151J135 Q09332
A0A1W4XG73 A0A1W4X5K4 U4U2M4 D6WKY8 N6UD09 A0A139WGR5 A0A1B6JT93 E0VWP7 A0A182GVT0 A0A1Y1LFV4 A0A2J7QLD5 A0A023EXD3 A0A1Q3FBA7 A0A1Q3FBF1 A0A1Q3FBB8 B0WIX0 A0A2S2QB88 Q17PC9 A0A1A9W760 A0A2M3Z0E1 A0A182QRW2 A0A182NSP3 A0A182LTU5 A0A2H8TN89 A0A2M4B8X7 A0A2M4B9I4 A0A182TQR4 A0A232F7H1 A0A1B0B6V6 A0A1A9Y0W5 A0A182UZT9 A0A2P8YV46 Q7QAS6 A0A182KXF2 A0A182HZ93 A0A084VDY2 A0A1B0CKK3 A0A1B6BXN7 A0A2M3YZF2 A0A182Y7E9 A0A1B0AI77 A0A1A9V3T6 A0A182WVU5 W4VRL2 A0A1B0G8K0 K7IP78 T1PM16 A0A2M3ZZJ0 A0A182RZF1 A0A0L0C6E8 A0A2M3ZZJ9 A0A182F6N1 A0A182J6H8 A0A336KG93 W5JE34 A0A1I8PAU5 A0A034W5E6 E2AEM6 A0A1L8E268 A0A3L8DM42 A0A1L8E1R7 J9JSC5 A0A2K8JM31 A0A0K8V5M4 A0A182JZ54 A0A0M8ZQZ6 A0A0P4VK86 B4QPX4 A0A1B0DJK7 F4X2J1 A0A0J9RXZ5 A0A3B0JS60 A0A088AHX7 A0A158NV88 A0A151WG09 A0A182TAR5 B4IFW7 A0A182PH54 Q2M0E2 A0A195FXM8 B4N4V0 A0A0M5IYM8 A0A023F3E2 A0A0A1XG58 A0A224XGM8 B4IYE1 A0A1W4UQX1 W8AWU6 A0A151J135 Q09332
EC Number
2.4.1.-
Pubmed
26354079
22118469
23537049
18362917
19820115
20566863
+ More
26483478 28004739 24945155 17510324 28648823 29403074 12364791 14747013 17210077 20966253 24438588 25244985 20075255 25315136 26108605 20920257 23761445 25348373 20798317 30249741 27129103 17994087 21719571 22936249 21347285 15632085 25474469 25830018 24495485 7729408 10731132 12537572 12537569
26483478 28004739 24945155 17510324 28648823 29403074 12364791 14747013 17210077 20966253 24438588 25244985 20075255 25315136 26108605 20920257 23761445 25348373 20798317 30249741 27129103 17994087 21719571 22936249 21347285 15632085 25474469 25830018 24495485 7729408 10731132 12537572 12537569
EMBL
NWSH01000353
PCG77109.1
RSAL01000040
RVE50987.1
KQ459602
KPI93423.1
+ More
AGBW02008329 OWR53644.1 KQ460615 KPJ13675.1 GALX01003280 JAB65186.1 KB631930 ERL87312.1 KQ971343 EFA03542.1 APGK01028346 APGK01028347 APGK01028348 KB740686 ENN79555.1 KYB27153.1 GECU01005291 JAT02416.1 DS235823 EEB17803.1 JXUM01002514 KQ560140 KXJ84210.1 GEZM01060864 JAV70845.1 NEVH01013247 PNF29392.1 GAPW01000017 JAC13581.1 GFDL01010181 JAV24864.1 GFDL01010203 JAV24842.1 GFDL01010195 JAV24850.1 DS231953 EDS28796.1 GGMS01005698 MBY74901.1 CH477193 EAT48502.1 GGFM01001200 MBW21951.1 AXCN02000232 AXCM01002283 GFXV01003828 MBW15633.1 GGFJ01000346 MBW49487.1 GGFJ01000347 MBW49488.1 NNAY01000809 OXU26398.1 JXJN01009254 PYGN01000342 PSN48119.1 AAAB01008888 EAA08752.5 APCN01005504 ATLV01011921 ATLV01011922 KE524735 KFB36176.1 AJWK01016363 AJWK01016364 AJWK01016365 AJWK01016366 AJWK01016367 GEDC01031519 GEDC01023173 JAS05779.1 JAS14125.1 GGFM01000899 MBW21650.1 GANO01002072 JAB57799.1 CCAG010000259 KA649105 KA649800 AFP63734.1 GGFK01000663 MBW33984.1 JRES01000836 KNC27850.1 GGFK01000673 MBW33994.1 UFQS01000429 UFQT01000429 SSX03864.1 SSX24229.1 ADMH02001795 ETN61074.1 GAKP01009436 GAKP01009432 GAKP01009430 JAC49522.1 GL438870 EFN68101.1 GFDF01001412 JAV12672.1 QOIP01000006 RLU21252.1 GFDF01001411 JAV12673.1 ABLF02034488 ABLF02034494 KY031137 ATU82888.1 GDHF01018133 JAI34181.1 KQ435969 KOX67853.1 GDKW01003694 JAI52901.1 CM000363 EDX11002.1 AJVK01065441 GL888584 EGI59316.1 CM002912 KMZ00458.1 OUUW01000002 SPP76196.1 ADTU01027005 ADTU01027006 KQ983203 KYQ46715.1 CH480834 EDW46554.1 CH379069 EAL30992.2 KQ981193 KYN45181.1 CH964101 EDW79389.1 CP012525 ALC43831.1 GBBI01002951 JAC15761.1 GBXI01004799 JAD09493.1 GFTR01008816 JAW07610.1 CH916366 EDV97614.1 GAMC01021269 JAB85286.1 KQ980575 KYN15496.1 U20554 AE014296 AY058353 BT044158
AGBW02008329 OWR53644.1 KQ460615 KPJ13675.1 GALX01003280 JAB65186.1 KB631930 ERL87312.1 KQ971343 EFA03542.1 APGK01028346 APGK01028347 APGK01028348 KB740686 ENN79555.1 KYB27153.1 GECU01005291 JAT02416.1 DS235823 EEB17803.1 JXUM01002514 KQ560140 KXJ84210.1 GEZM01060864 JAV70845.1 NEVH01013247 PNF29392.1 GAPW01000017 JAC13581.1 GFDL01010181 JAV24864.1 GFDL01010203 JAV24842.1 GFDL01010195 JAV24850.1 DS231953 EDS28796.1 GGMS01005698 MBY74901.1 CH477193 EAT48502.1 GGFM01001200 MBW21951.1 AXCN02000232 AXCM01002283 GFXV01003828 MBW15633.1 GGFJ01000346 MBW49487.1 GGFJ01000347 MBW49488.1 NNAY01000809 OXU26398.1 JXJN01009254 PYGN01000342 PSN48119.1 AAAB01008888 EAA08752.5 APCN01005504 ATLV01011921 ATLV01011922 KE524735 KFB36176.1 AJWK01016363 AJWK01016364 AJWK01016365 AJWK01016366 AJWK01016367 GEDC01031519 GEDC01023173 JAS05779.1 JAS14125.1 GGFM01000899 MBW21650.1 GANO01002072 JAB57799.1 CCAG010000259 KA649105 KA649800 AFP63734.1 GGFK01000663 MBW33984.1 JRES01000836 KNC27850.1 GGFK01000673 MBW33994.1 UFQS01000429 UFQT01000429 SSX03864.1 SSX24229.1 ADMH02001795 ETN61074.1 GAKP01009436 GAKP01009432 GAKP01009430 JAC49522.1 GL438870 EFN68101.1 GFDF01001412 JAV12672.1 QOIP01000006 RLU21252.1 GFDF01001411 JAV12673.1 ABLF02034488 ABLF02034494 KY031137 ATU82888.1 GDHF01018133 JAI34181.1 KQ435969 KOX67853.1 GDKW01003694 JAI52901.1 CM000363 EDX11002.1 AJVK01065441 GL888584 EGI59316.1 CM002912 KMZ00458.1 OUUW01000002 SPP76196.1 ADTU01027005 ADTU01027006 KQ983203 KYQ46715.1 CH480834 EDW46554.1 CH379069 EAL30992.2 KQ981193 KYN45181.1 CH964101 EDW79389.1 CP012525 ALC43831.1 GBBI01002951 JAC15761.1 GBXI01004799 JAD09493.1 GFTR01008816 JAW07610.1 CH916366 EDV97614.1 GAMC01021269 JAB85286.1 KQ980575 KYN15496.1 U20554 AE014296 AY058353 BT044158
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000192223
+ More
UP000030742 UP000007266 UP000019118 UP000009046 UP000069940 UP000249989 UP000235965 UP000002320 UP000008820 UP000091820 UP000075886 UP000075884 UP000075883 UP000075902 UP000215335 UP000092460 UP000092443 UP000075903 UP000245037 UP000007062 UP000075882 UP000075840 UP000030765 UP000092461 UP000076408 UP000092445 UP000078200 UP000076407 UP000092444 UP000002358 UP000095301 UP000075900 UP000037069 UP000069272 UP000075880 UP000000673 UP000095300 UP000000311 UP000279307 UP000007819 UP000075881 UP000053105 UP000000304 UP000092462 UP000007755 UP000268350 UP000005203 UP000005205 UP000075809 UP000075901 UP000001292 UP000075885 UP000001819 UP000078541 UP000007798 UP000092553 UP000001070 UP000192221 UP000078492 UP000000803
UP000030742 UP000007266 UP000019118 UP000009046 UP000069940 UP000249989 UP000235965 UP000002320 UP000008820 UP000091820 UP000075886 UP000075884 UP000075883 UP000075902 UP000215335 UP000092460 UP000092443 UP000075903 UP000245037 UP000007062 UP000075882 UP000075840 UP000030765 UP000092461 UP000076408 UP000092445 UP000078200 UP000076407 UP000092444 UP000002358 UP000095301 UP000075900 UP000037069 UP000069272 UP000075880 UP000000673 UP000095300 UP000000311 UP000279307 UP000007819 UP000075881 UP000053105 UP000000304 UP000092462 UP000007755 UP000268350 UP000005203 UP000005205 UP000075809 UP000075901 UP000001292 UP000075885 UP000001819 UP000078541 UP000007798 UP000092553 UP000001070 UP000192221 UP000078492 UP000000803
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JZ71
A0A3S2PGS0
A0A194PK85
A0A212FIU0
A0A194R8U7
V5I9A4
+ More
A0A1W4XG73 A0A1W4X5K4 U4U2M4 D6WKY8 N6UD09 A0A139WGR5 A0A1B6JT93 E0VWP7 A0A182GVT0 A0A1Y1LFV4 A0A2J7QLD5 A0A023EXD3 A0A1Q3FBA7 A0A1Q3FBF1 A0A1Q3FBB8 B0WIX0 A0A2S2QB88 Q17PC9 A0A1A9W760 A0A2M3Z0E1 A0A182QRW2 A0A182NSP3 A0A182LTU5 A0A2H8TN89 A0A2M4B8X7 A0A2M4B9I4 A0A182TQR4 A0A232F7H1 A0A1B0B6V6 A0A1A9Y0W5 A0A182UZT9 A0A2P8YV46 Q7QAS6 A0A182KXF2 A0A182HZ93 A0A084VDY2 A0A1B0CKK3 A0A1B6BXN7 A0A2M3YZF2 A0A182Y7E9 A0A1B0AI77 A0A1A9V3T6 A0A182WVU5 W4VRL2 A0A1B0G8K0 K7IP78 T1PM16 A0A2M3ZZJ0 A0A182RZF1 A0A0L0C6E8 A0A2M3ZZJ9 A0A182F6N1 A0A182J6H8 A0A336KG93 W5JE34 A0A1I8PAU5 A0A034W5E6 E2AEM6 A0A1L8E268 A0A3L8DM42 A0A1L8E1R7 J9JSC5 A0A2K8JM31 A0A0K8V5M4 A0A182JZ54 A0A0M8ZQZ6 A0A0P4VK86 B4QPX4 A0A1B0DJK7 F4X2J1 A0A0J9RXZ5 A0A3B0JS60 A0A088AHX7 A0A158NV88 A0A151WG09 A0A182TAR5 B4IFW7 A0A182PH54 Q2M0E2 A0A195FXM8 B4N4V0 A0A0M5IYM8 A0A023F3E2 A0A0A1XG58 A0A224XGM8 B4IYE1 A0A1W4UQX1 W8AWU6 A0A151J135 Q09332
A0A1W4XG73 A0A1W4X5K4 U4U2M4 D6WKY8 N6UD09 A0A139WGR5 A0A1B6JT93 E0VWP7 A0A182GVT0 A0A1Y1LFV4 A0A2J7QLD5 A0A023EXD3 A0A1Q3FBA7 A0A1Q3FBF1 A0A1Q3FBB8 B0WIX0 A0A2S2QB88 Q17PC9 A0A1A9W760 A0A2M3Z0E1 A0A182QRW2 A0A182NSP3 A0A182LTU5 A0A2H8TN89 A0A2M4B8X7 A0A2M4B9I4 A0A182TQR4 A0A232F7H1 A0A1B0B6V6 A0A1A9Y0W5 A0A182UZT9 A0A2P8YV46 Q7QAS6 A0A182KXF2 A0A182HZ93 A0A084VDY2 A0A1B0CKK3 A0A1B6BXN7 A0A2M3YZF2 A0A182Y7E9 A0A1B0AI77 A0A1A9V3T6 A0A182WVU5 W4VRL2 A0A1B0G8K0 K7IP78 T1PM16 A0A2M3ZZJ0 A0A182RZF1 A0A0L0C6E8 A0A2M3ZZJ9 A0A182F6N1 A0A182J6H8 A0A336KG93 W5JE34 A0A1I8PAU5 A0A034W5E6 E2AEM6 A0A1L8E268 A0A3L8DM42 A0A1L8E1R7 J9JSC5 A0A2K8JM31 A0A0K8V5M4 A0A182JZ54 A0A0M8ZQZ6 A0A0P4VK86 B4QPX4 A0A1B0DJK7 F4X2J1 A0A0J9RXZ5 A0A3B0JS60 A0A088AHX7 A0A158NV88 A0A151WG09 A0A182TAR5 B4IFW7 A0A182PH54 Q2M0E2 A0A195FXM8 B4N4V0 A0A0M5IYM8 A0A023F3E2 A0A0A1XG58 A0A224XGM8 B4IYE1 A0A1W4UQX1 W8AWU6 A0A151J135 Q09332
PDB
5Y7O
E-value=1.21772e-25,
Score=292
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum lumen
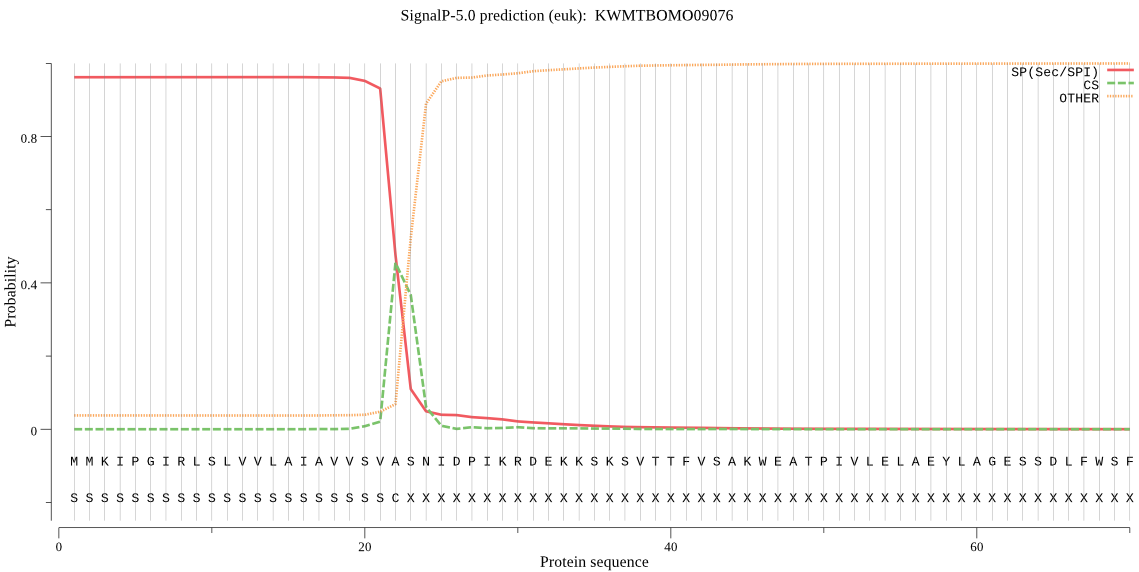
SignalP
Position: 1 - 22,
Likelihood: 0.961822
Length:
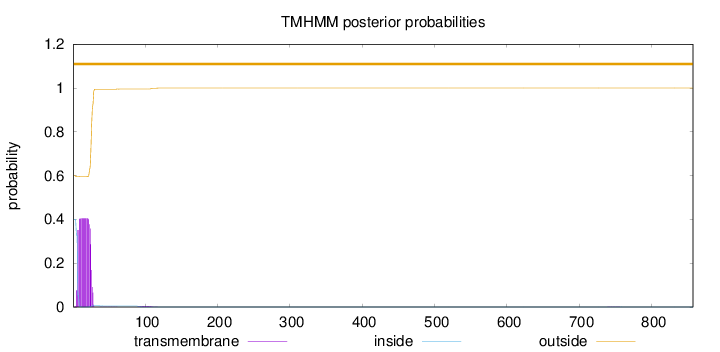
857
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.97509999999998
Exp number, first 60 AAs:
7.88714
Total prob of N-in:
0.39970
outside
1 - 857
Population Genetic Test Statistics
Pi
313.948407
Theta
203.168455
Tajima's D
1.578248
CLR
0.012337
CSRT
0.8025598720064
Interpretation
Uncertain
