Pre Gene Modal
BGIBMGA007509
Annotation
PREDICTED:_beta-catenin-like_protein_1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.483
Sequence
CDS
ATGGATGTTGCCGAGCTATTGACATTCAAACCTACGCCAACACCAAAACGGCCAAATGAAGATGAGAGCGATGAGGAGGATCAAGCCAAAAGTGCTAAAATTCGTCGTACCACCAAACTTAAACAGGAACCTGTACATTTGAGCAAGAATATGTTGAAGGAACCCTCGATTACTGATAAAGAACGAGAAGATATTCTTAAATTTGTTGCCACTGAAGTAACAGAGGGAGAAGTACTAGATGAAACCACAGTGAAGAAACTAGTTCTCAATTTTGAAAAGAAAGCACTTAGAAATAGAGAGATGAGGATTAAATTTCCTGATCAGCCAGACAAGTTTATGGACAGTGAAATTGATTTACATGATGCATTACAGGATTTAAGTGCTGTCGCAACAGTACCAGACCAGTATCCTCTGCTTGTGGAATTAAAGTGTGTGAACTCATTATTAGAATTACTATCACATGATAATACTGATGTCTCCACCAAAGTTGTTCAACTGCTACAGGAAATGACAGATGTTGACATACTCCATGAGAGTGAGGAAGGTGCGGAAGATTTAATAAATGCAATGGCTGAAGCTGAAGCACCTGCATTATTACTACACAACCTGACTCGATTGGATGAGCAGGTACCAGATGAACGAGACGCTGTCCATAATACACTGGGTATTATTGAAAATATCACAGAATTCAGACCAGAGCTTTGTATTGAAGTAGCAAAACAAGGATTTTTACAGTGGATTCTAAAAAGACTGAAGGTTAAACTGCCTTTTGATGGGAACAAACTGTATGCAACAGAAATTTTATCTATATTACTTCAGAACACTCCCGAAAACAGAAAACTACTGGGAGAACTTGATGGCATTGATGTTTTACTACAACAGCTAGCGTTTTACAAAAGACACGACCCCAGCGGCGCCGAAGAACAAGAGGCGATGGAGAACATGTTCGACTCACTGTGCTGCGCGCTCATGGAGCCCTCCAACCGGGACCGCTTCCTGCGCGGCGAGGGACTGCAACTCATGAACCTCATGTTGAGAGAGAAAAAAATGTCACGAAATGGTTCTTTGAAAGTACTGGATCACGCTCTCGTGGGACCCGACGGTCGCGATAATTGCAATAAATTTGTAGATATACTGGGGCTCCGAACTGTGTTTCCACTTTTTATGAAAACACCCAAAAGAAAACGATTATTAACTGTTGATCAACACGAAGAGCACGTAGTATCAATAATAGCATCCATGTTACGAAACTGTCAGGGAAGTCAAAGGCAGAGACTGTTGGCGAAGTTCACTGAAAACGATCTAGAGAAAGTAGATCGGTTGTTGGAGCTACATTTCAAATACATGGACAAAGTCGACAGAACCGAAAAAGATATGGAGCGTGAAACTGAAGAATTAGATGATGACGCTCAGTATTTGAAAAGGCTCTCGGGCGGTTTATTCACATTGCAACTTATAGACAGAATAATTTTGGAGGTGTGTACGGCCGGTCCGTCGACGATAAAGCAACGCGTCCAGCGTGTGTTGTCATTACGAGGCGGCTCCCTCAAGATTATCAGACATGTAATGAGGGAATATGCGGGCAACCTCGGCGACGCTGGAAGCGAGGAGTGGCGGCAACAAGAACAACAGCATATATTGCAACTCGTTGATAAGTTTTAA
Protein
MDVAELLTFKPTPTPKRPNEDESDEEDQAKSAKIRRTTKLKQEPVHLSKNMLKEPSITDKEREDILKFVATEVTEGEVLDETTVKKLVLNFEKKALRNREMRIKFPDQPDKFMDSEIDLHDALQDLSAVATVPDQYPLLVELKCVNSLLELLSHDNTDVSTKVVQLLQEMTDVDILHESEEGAEDLINAMAEAEAPALLLHNLTRLDEQVPDERDAVHNTLGIIENITEFRPELCIEVAKQGFLQWILKRLKVKLPFDGNKLYATEILSILLQNTPENRKLLGELDGIDVLLQQLAFYKRHDPSGAEEQEAMENMFDSLCCALMEPSNRDRFLRGEGLQLMNLMLREKKMSRNGSLKVLDHALVGPDGRDNCNKFVDILGLRTVFPLFMKTPKRKRLLTVDQHEEHVVSIIASMLRNCQGSQRQRLLAKFTENDLEKVDRLLELHFKYMDKVDRTEKDMERETEELDDDAQYLKRLSGGLFTLQLIDRIILEVCTAGPSTIKQRVQRVLSLRGGSLKIIRHVMREYAGNLGDAGSEEWRQQEQQHILQLVDKF
Summary
Uniprot
H9JDB2
A0A194R7P2
A0A194PL10
A0A2A4JI02
A0A212FIT2
A0A437BL22
+ More
A0A2H1WDF2 A0A0L7LDC9 A0A1Y1MC37 D7EK03 A0A1W4WKE4 A0A0M9A5U4 A0A088AM02 Q17P30 A0A0C9QIS2 A0A158N981 F4WM13 A0A0T6B1G0 A0A195FTV9 A0A0L7RIC5 A0A1B6FUB5 A0A1B6IAI5 E2C2K6 A0A1B6L8U3 N6T0L4 A0A182GQG4 A0A195DCL7 A0A023EWI4 A0A195BC52 A0A0J7KE12 A0A182GF37 A0A026W247 B0WFG5 A0A182VDC2 A0A067R1X8 A0A182P2K2 A0A195C640 A0A2M4APL1 A0A182L8X6 Q7PXD5 A0A182Q491 A0A182X9Q0 A0A2A3EE28 A0A0A9XX76 A0A151X9X8 A0A182TIR3 A0A182HKN5 W5JTW5 A0A182N491 A0A182VR60 A0A182F755 A0A1Q3F931 A0A182RZ84 A0A182YGC5 A0A2J7RHJ0 A0A182M1E8 A0A182KG51 A0A0P4VR56 A0A2R7VUI4 A0A2M4BIX0 U4U114 A0A069DVH7 A0A1B6C874 A0A0L0CLF8 A0A0V0G9B5 J9K512 W8BU53 A0A2H8TSC4 B4HKY9 A0A034VQ25 Q9VHJ7 A0A0K8VPS6 A0A1I8MJ47 A0A1A9XQ01 A0A1B0C345 T1PIW9 A0A154P4S7 A0A1I8NZ44 A0A0A1X6S5 A0A1B0G6Z2 A0A1A9V8I2 A0A1B0A9J4 B4N818 A0A1L8DWX8 A0A1S4EMH2 B4K9E7 A0A182STN5 A0A1L8DWG5 A0A1B0D9P9 B4QXC6 B4PTX8 B4LXK8 Q295Q5 K7IWE7 A0A3B0KH54 T1HUZ2 A0A3B0JVJ7 B3P1W2 B4JTJ4 B4GFU7
A0A2H1WDF2 A0A0L7LDC9 A0A1Y1MC37 D7EK03 A0A1W4WKE4 A0A0M9A5U4 A0A088AM02 Q17P30 A0A0C9QIS2 A0A158N981 F4WM13 A0A0T6B1G0 A0A195FTV9 A0A0L7RIC5 A0A1B6FUB5 A0A1B6IAI5 E2C2K6 A0A1B6L8U3 N6T0L4 A0A182GQG4 A0A195DCL7 A0A023EWI4 A0A195BC52 A0A0J7KE12 A0A182GF37 A0A026W247 B0WFG5 A0A182VDC2 A0A067R1X8 A0A182P2K2 A0A195C640 A0A2M4APL1 A0A182L8X6 Q7PXD5 A0A182Q491 A0A182X9Q0 A0A2A3EE28 A0A0A9XX76 A0A151X9X8 A0A182TIR3 A0A182HKN5 W5JTW5 A0A182N491 A0A182VR60 A0A182F755 A0A1Q3F931 A0A182RZ84 A0A182YGC5 A0A2J7RHJ0 A0A182M1E8 A0A182KG51 A0A0P4VR56 A0A2R7VUI4 A0A2M4BIX0 U4U114 A0A069DVH7 A0A1B6C874 A0A0L0CLF8 A0A0V0G9B5 J9K512 W8BU53 A0A2H8TSC4 B4HKY9 A0A034VQ25 Q9VHJ7 A0A0K8VPS6 A0A1I8MJ47 A0A1A9XQ01 A0A1B0C345 T1PIW9 A0A154P4S7 A0A1I8NZ44 A0A0A1X6S5 A0A1B0G6Z2 A0A1A9V8I2 A0A1B0A9J4 B4N818 A0A1L8DWX8 A0A1S4EMH2 B4K9E7 A0A182STN5 A0A1L8DWG5 A0A1B0D9P9 B4QXC6 B4PTX8 B4LXK8 Q295Q5 K7IWE7 A0A3B0KH54 T1HUZ2 A0A3B0JVJ7 B3P1W2 B4JTJ4 B4GFU7
Pubmed
19121390
26354079
22118469
26227816
28004739
18362917
+ More
19820115 17510324 21347285 21719571 20798317 23537049 26483478 24945155 24508170 30249741 24845553 20966253 12364791 14747013 17210077 25401762 26823975 20920257 23761445 25244985 27129103 26334808 26108605 24495485 17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 17550304 15632085 20075255
19820115 17510324 21347285 21719571 20798317 23537049 26483478 24945155 24508170 30249741 24845553 20966253 12364791 14747013 17210077 25401762 26823975 20920257 23761445 25244985 27129103 26334808 26108605 24495485 17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 25830018 17550304 15632085 20075255
EMBL
BABH01030150
BABH01030151
KQ460615
KPJ13667.1
KQ459602
KPI93414.1
+ More
NWSH01001498 PCG71063.1 AGBW02008329 OWR53653.1 RSAL01000040 RVE50979.1 ODYU01007888 SOQ51057.1 JTDY01001602 KOB73404.1 GEZM01037132 JAV82150.1 DS497724 EFA12950.1 KQ435727 KOX77840.1 CH477194 EAT48471.1 GBYB01014638 GBYB01014639 JAG84405.1 JAG84406.1 ADTU01001367 GL888217 EGI64657.1 LJIG01016201 KRT81311.1 KQ981264 KYN44055.1 KQ414584 KOC70584.1 GECZ01015994 GECZ01005817 JAS53775.1 JAS63952.1 GECU01029976 GECU01023778 JAS77730.1 JAS83928.1 GL452135 EFN77862.1 GEBQ01019946 JAT20031.1 APGK01057248 KB741279 ENN71043.1 JXUM01015742 KQ560438 KXJ82411.1 KQ980989 KYN10628.1 GAPW01000744 JAC12854.1 KQ976519 KYM82128.1 LBMM01008686 KMQ88698.1 JXUM01059025 KQ562031 KXJ76861.1 KK107474 QOIP01000002 EZA50155.1 RLU25397.1 DS231918 EDS26233.1 KK853083 KDR11648.1 KQ978220 KYM96329.1 GGFK01009415 MBW42736.1 AAAB01008987 EAA01129.1 AXCN02000029 KZ288282 PBC29542.1 GBHO01019613 GBRD01000859 GDHC01003388 JAG23991.1 JAG64962.1 JAQ15241.1 KQ982353 KYQ57177.1 APCN01000853 ADMH02000111 ETN67797.1 GFDL01011022 JAV24023.1 NEVH01003741 PNF40287.1 AXCM01011710 GDKW01001088 JAI55507.1 KK854093 PTY11156.1 GGFJ01003868 MBW53009.1 KB631120 KB631121 KB631861 ERL84120.1 ERL84123.1 ERL86767.1 GBGD01001009 JAC87880.1 GEDC01027600 JAS09698.1 JRES01000223 KNC33203.1 GECL01001696 JAP04428.1 ABLF02010177 GAMC01006102 JAC00454.1 GFXV01005311 MBW17116.1 CH480815 EDW42953.1 GAKP01014380 GAKP01014378 JAC44574.1 AE014297 AY051943 AAF54309.1 AAK93367.1 GDHF01011425 JAI40889.1 JXJN01024780 KA648711 AFP63340.1 KQ434806 KZC06120.1 GBXI01009437 GBXI01007681 JAD04855.1 JAD06611.1 CCAG010018274 CH964232 EDW81269.1 GFDF01003309 JAV10775.1 CH933806 EDW15579.1 GFDF01003308 JAV10776.1 AJVK01013016 CM000364 EDX13697.1 CM000160 EDW96589.1 CH940650 EDW66792.1 CM000070 EAL28653.1 OUUW01000008 SPP84431.1 ACPB03002116 SPP84432.1 CH954181 EDV49850.1 CH916373 EDV95084.1 CH479182 EDW34482.1
NWSH01001498 PCG71063.1 AGBW02008329 OWR53653.1 RSAL01000040 RVE50979.1 ODYU01007888 SOQ51057.1 JTDY01001602 KOB73404.1 GEZM01037132 JAV82150.1 DS497724 EFA12950.1 KQ435727 KOX77840.1 CH477194 EAT48471.1 GBYB01014638 GBYB01014639 JAG84405.1 JAG84406.1 ADTU01001367 GL888217 EGI64657.1 LJIG01016201 KRT81311.1 KQ981264 KYN44055.1 KQ414584 KOC70584.1 GECZ01015994 GECZ01005817 JAS53775.1 JAS63952.1 GECU01029976 GECU01023778 JAS77730.1 JAS83928.1 GL452135 EFN77862.1 GEBQ01019946 JAT20031.1 APGK01057248 KB741279 ENN71043.1 JXUM01015742 KQ560438 KXJ82411.1 KQ980989 KYN10628.1 GAPW01000744 JAC12854.1 KQ976519 KYM82128.1 LBMM01008686 KMQ88698.1 JXUM01059025 KQ562031 KXJ76861.1 KK107474 QOIP01000002 EZA50155.1 RLU25397.1 DS231918 EDS26233.1 KK853083 KDR11648.1 KQ978220 KYM96329.1 GGFK01009415 MBW42736.1 AAAB01008987 EAA01129.1 AXCN02000029 KZ288282 PBC29542.1 GBHO01019613 GBRD01000859 GDHC01003388 JAG23991.1 JAG64962.1 JAQ15241.1 KQ982353 KYQ57177.1 APCN01000853 ADMH02000111 ETN67797.1 GFDL01011022 JAV24023.1 NEVH01003741 PNF40287.1 AXCM01011710 GDKW01001088 JAI55507.1 KK854093 PTY11156.1 GGFJ01003868 MBW53009.1 KB631120 KB631121 KB631861 ERL84120.1 ERL84123.1 ERL86767.1 GBGD01001009 JAC87880.1 GEDC01027600 JAS09698.1 JRES01000223 KNC33203.1 GECL01001696 JAP04428.1 ABLF02010177 GAMC01006102 JAC00454.1 GFXV01005311 MBW17116.1 CH480815 EDW42953.1 GAKP01014380 GAKP01014378 JAC44574.1 AE014297 AY051943 AAF54309.1 AAK93367.1 GDHF01011425 JAI40889.1 JXJN01024780 KA648711 AFP63340.1 KQ434806 KZC06120.1 GBXI01009437 GBXI01007681 JAD04855.1 JAD06611.1 CCAG010018274 CH964232 EDW81269.1 GFDF01003309 JAV10775.1 CH933806 EDW15579.1 GFDF01003308 JAV10776.1 AJVK01013016 CM000364 EDX13697.1 CM000160 EDW96589.1 CH940650 EDW66792.1 CM000070 EAL28653.1 OUUW01000008 SPP84431.1 ACPB03002116 SPP84432.1 CH954181 EDV49850.1 CH916373 EDV95084.1 CH479182 EDW34482.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
+ More
UP000037510 UP000007266 UP000192223 UP000053105 UP000005203 UP000008820 UP000005205 UP000007755 UP000078541 UP000053825 UP000008237 UP000019118 UP000069940 UP000249989 UP000078492 UP000078540 UP000036403 UP000053097 UP000279307 UP000002320 UP000075903 UP000027135 UP000075885 UP000078542 UP000075882 UP000007062 UP000075886 UP000076407 UP000242457 UP000075809 UP000075902 UP000075840 UP000000673 UP000075884 UP000075920 UP000069272 UP000075900 UP000076408 UP000235965 UP000075883 UP000075881 UP000030742 UP000037069 UP000007819 UP000001292 UP000000803 UP000095301 UP000092443 UP000092460 UP000076502 UP000095300 UP000092444 UP000078200 UP000092445 UP000007798 UP000079169 UP000009192 UP000075901 UP000092462 UP000000304 UP000002282 UP000008792 UP000001819 UP000002358 UP000268350 UP000015103 UP000008711 UP000001070 UP000008744
UP000037510 UP000007266 UP000192223 UP000053105 UP000005203 UP000008820 UP000005205 UP000007755 UP000078541 UP000053825 UP000008237 UP000019118 UP000069940 UP000249989 UP000078492 UP000078540 UP000036403 UP000053097 UP000279307 UP000002320 UP000075903 UP000027135 UP000075885 UP000078542 UP000075882 UP000007062 UP000075886 UP000076407 UP000242457 UP000075809 UP000075902 UP000075840 UP000000673 UP000075884 UP000075920 UP000069272 UP000075900 UP000076408 UP000235965 UP000075883 UP000075881 UP000030742 UP000037069 UP000007819 UP000001292 UP000000803 UP000095301 UP000092443 UP000092460 UP000076502 UP000095300 UP000092444 UP000078200 UP000092445 UP000007798 UP000079169 UP000009192 UP000075901 UP000092462 UP000000304 UP000002282 UP000008792 UP000001819 UP000002358 UP000268350 UP000015103 UP000008711 UP000001070 UP000008744
Pfam
PF08216 CTNNBL
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JDB2
A0A194R7P2
A0A194PL10
A0A2A4JI02
A0A212FIT2
A0A437BL22
+ More
A0A2H1WDF2 A0A0L7LDC9 A0A1Y1MC37 D7EK03 A0A1W4WKE4 A0A0M9A5U4 A0A088AM02 Q17P30 A0A0C9QIS2 A0A158N981 F4WM13 A0A0T6B1G0 A0A195FTV9 A0A0L7RIC5 A0A1B6FUB5 A0A1B6IAI5 E2C2K6 A0A1B6L8U3 N6T0L4 A0A182GQG4 A0A195DCL7 A0A023EWI4 A0A195BC52 A0A0J7KE12 A0A182GF37 A0A026W247 B0WFG5 A0A182VDC2 A0A067R1X8 A0A182P2K2 A0A195C640 A0A2M4APL1 A0A182L8X6 Q7PXD5 A0A182Q491 A0A182X9Q0 A0A2A3EE28 A0A0A9XX76 A0A151X9X8 A0A182TIR3 A0A182HKN5 W5JTW5 A0A182N491 A0A182VR60 A0A182F755 A0A1Q3F931 A0A182RZ84 A0A182YGC5 A0A2J7RHJ0 A0A182M1E8 A0A182KG51 A0A0P4VR56 A0A2R7VUI4 A0A2M4BIX0 U4U114 A0A069DVH7 A0A1B6C874 A0A0L0CLF8 A0A0V0G9B5 J9K512 W8BU53 A0A2H8TSC4 B4HKY9 A0A034VQ25 Q9VHJ7 A0A0K8VPS6 A0A1I8MJ47 A0A1A9XQ01 A0A1B0C345 T1PIW9 A0A154P4S7 A0A1I8NZ44 A0A0A1X6S5 A0A1B0G6Z2 A0A1A9V8I2 A0A1B0A9J4 B4N818 A0A1L8DWX8 A0A1S4EMH2 B4K9E7 A0A182STN5 A0A1L8DWG5 A0A1B0D9P9 B4QXC6 B4PTX8 B4LXK8 Q295Q5 K7IWE7 A0A3B0KH54 T1HUZ2 A0A3B0JVJ7 B3P1W2 B4JTJ4 B4GFU7
A0A2H1WDF2 A0A0L7LDC9 A0A1Y1MC37 D7EK03 A0A1W4WKE4 A0A0M9A5U4 A0A088AM02 Q17P30 A0A0C9QIS2 A0A158N981 F4WM13 A0A0T6B1G0 A0A195FTV9 A0A0L7RIC5 A0A1B6FUB5 A0A1B6IAI5 E2C2K6 A0A1B6L8U3 N6T0L4 A0A182GQG4 A0A195DCL7 A0A023EWI4 A0A195BC52 A0A0J7KE12 A0A182GF37 A0A026W247 B0WFG5 A0A182VDC2 A0A067R1X8 A0A182P2K2 A0A195C640 A0A2M4APL1 A0A182L8X6 Q7PXD5 A0A182Q491 A0A182X9Q0 A0A2A3EE28 A0A0A9XX76 A0A151X9X8 A0A182TIR3 A0A182HKN5 W5JTW5 A0A182N491 A0A182VR60 A0A182F755 A0A1Q3F931 A0A182RZ84 A0A182YGC5 A0A2J7RHJ0 A0A182M1E8 A0A182KG51 A0A0P4VR56 A0A2R7VUI4 A0A2M4BIX0 U4U114 A0A069DVH7 A0A1B6C874 A0A0L0CLF8 A0A0V0G9B5 J9K512 W8BU53 A0A2H8TSC4 B4HKY9 A0A034VQ25 Q9VHJ7 A0A0K8VPS6 A0A1I8MJ47 A0A1A9XQ01 A0A1B0C345 T1PIW9 A0A154P4S7 A0A1I8NZ44 A0A0A1X6S5 A0A1B0G6Z2 A0A1A9V8I2 A0A1B0A9J4 B4N818 A0A1L8DWX8 A0A1S4EMH2 B4K9E7 A0A182STN5 A0A1L8DWG5 A0A1B0D9P9 B4QXC6 B4PTX8 B4LXK8 Q295Q5 K7IWE7 A0A3B0KH54 T1HUZ2 A0A3B0JVJ7 B3P1W2 B4JTJ4 B4GFU7
PDB
4HM9
E-value=1.26491e-159,
Score=1446
Ontologies
GO
PANTHER
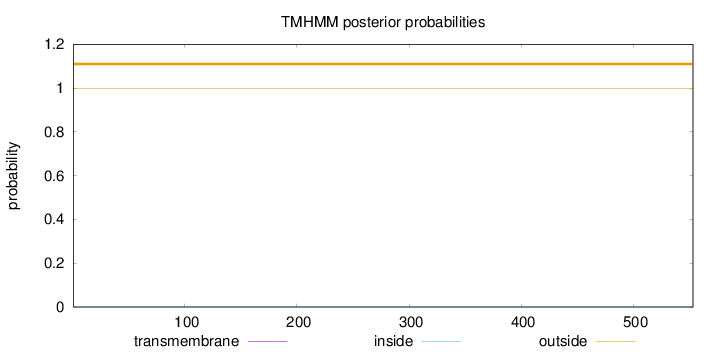
Topology
Length:
553
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00049
outside
1 - 553
Population Genetic Test Statistics
Pi
260.820833
Theta
194.175563
Tajima's D
1.658202
CLR
0.438297
CSRT
0.817859107044648
Interpretation
Uncertain
