Pre Gene Modal
BGIBMGA007507
Annotation
PREDICTED:_probable_ubiquitin_carboxyl-terminal_hydrolase_FAF-X_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.767 Nuclear Reliability : 2.095
Sequence
CDS
ATGAAAGAACAAGACCAGATGCCTGGGGAGATGGCATTCCCTGAGGCGAAATTAAAGGCACTGGAAGACAAACTTTCACATCCCCGTTGGGTTGTACCAGTGCTGCCAGAACAAGAATTGGAAGCTCTTCTAGTCGCTGCAACAGAACTAGCTGCAAAAGGAGAAGATGTACACAATGCGGCGTGTCAGAAATTTTACACTGATGCTCTCACAATATCTTTTACAAAGATATTAACAGATGATGCAGTATCATCGTGGAAAAATAATATACAACAGTGTGTAAGATCCAACTGTGAGAAGCTAGTCAAGTTGTGTGCTCTTAAACTTGATGATCCCAGATTTTTGCATCTATTGACTATGACATTAAACCCCAGTAACAAGTTTCATATGTTCAATGCATCGCGAACGAGCGAGGGCCTTTCGGTGACATCAAATACCACTGGTACAGGTCAAGGGCCTTCTCAAGACATAGAAATTTTTGCTAGGTCTCATGATCATCGTACTCCTAGGGGGTGGCTGGTCCATCTAATTAATGTGTTTGGTCAAGCAGGCGGCTTTGTGAAATTAAGGGAGAGGTTTGAAGAGATAATGGGTTTTCAAAAGTCTGATCTCAATTCATCTATGAATACAGAAGAAAAGGTATTGTCTGAGATGATGGAAAAAGAAAGTGATGAGAGCAAGACATCTAACACCAATTGTATTGAACCAGTTCTTGTTATTGGGGAAAACAGTGATGTAAGTACTAATTTAAAATTATTTTAA
Protein
MKEQDQMPGEMAFPEAKLKALEDKLSHPRWVVPVLPEQELEALLVAATELAAKGEDVHNAACQKFYTDALTISFTKILTDDAVSSWKNNIQQCVRSNCEKLVKLCALKLDDPRFLHLLTMTLNPSNKFHMFNASRTSEGLSVTSNTTGTGQGPSQDIEIFARSHDHRTPRGWLVHLINVFGQAGGFVKLRERFEEIMGFQKSDLNSSMNTEEKVLSEMMEKESDESKTSNTNCIEPVLVIGENSDVSTNLKLF
Summary
Cofactor
FMN
Similarity
Belongs to the peptidase C19 family.
Belongs to the FMN-dependent alpha-hydroxy acid dehydrogenase family.
Belongs to the FMN-dependent alpha-hydroxy acid dehydrogenase family.
Uniprot
H9JDB0
A0A212EP08
A0A2A4J087
A0A2A4J0N2
A0A3S2TP78
A0A2H1WVS5
+ More
A0A194PL04 A0A194R833 A0A310SNR3 A0A2A3EAM0 A0A088A5Z8 A0A224XFS3 A0A1B6EYR0 A0A067RUD8 A0A069DY94 A0A2J7R122 A0A1B6HSU7 A0A2J7R116 A0A0J7NNW5 A0A1B6C8S8 A0A3L8DR29 A0A154P3H2 A0A0L7QM89 E2A416 A0A232F7U1 K7IRB1 A0A195C536 U4UCY8 N6TM76 D6WKR1 A0A139WI67 A0A151JRZ4 F4W771 A0A151WV29 A0A195BJS5 A0A158NJ55 A0A195EYM8 A0A0T6AU90 A0A1Y1LX88 A0A2D4PNU0 C3ZE43 A0A1E1XGF4 A0A443SJY2 A0A2R5LEU6 A0A433SLI9 B4DGX9 A0A2R2MQM1 A0A0N0BI91 A0A1S3IUS1 A0A2K6GZY0 E0VC08 A0A2P4TGZ1 A0A3Q2HLU3 A0A2U4C119 A0A2K6H024 A0A3Q2IDU3 A0A2U4C0Y1 F6WF49 A0A2Y9S8T1 L8IWX3 A0A2Y9S7Q2 A0A2Y9LTS1 A0A2U3V074 A0A2U4C0S9 A0A384ACM9 A0A2Y9LQT8 G5E630 A0A384ACG5 A0A452FL98 W5NVJ3 F7CE06 A0A146KRP5 A0A146LHV0 A0A2U4C0W6 A0A2Y9LYK2 G3TXD1 A0A2Y9S0S0 G3T223 U3JFP5 A0A287AC95 A0A287AM69 E9PWA9 Q8C7T5 A0A287A891 A0A287B4Y6 V4AR29 G3GSH2 A0A0N8CJH9 I3LMY9 A0A0P5ZXL4 A0A0P5U0S7 A0A0P6GU97 A0A0P5CYN1 A0A0P5AML0 A0A0P5VQM3 A0A0N8CJW0 A0A3L7H1V6
A0A194PL04 A0A194R833 A0A310SNR3 A0A2A3EAM0 A0A088A5Z8 A0A224XFS3 A0A1B6EYR0 A0A067RUD8 A0A069DY94 A0A2J7R122 A0A1B6HSU7 A0A2J7R116 A0A0J7NNW5 A0A1B6C8S8 A0A3L8DR29 A0A154P3H2 A0A0L7QM89 E2A416 A0A232F7U1 K7IRB1 A0A195C536 U4UCY8 N6TM76 D6WKR1 A0A139WI67 A0A151JRZ4 F4W771 A0A151WV29 A0A195BJS5 A0A158NJ55 A0A195EYM8 A0A0T6AU90 A0A1Y1LX88 A0A2D4PNU0 C3ZE43 A0A1E1XGF4 A0A443SJY2 A0A2R5LEU6 A0A433SLI9 B4DGX9 A0A2R2MQM1 A0A0N0BI91 A0A1S3IUS1 A0A2K6GZY0 E0VC08 A0A2P4TGZ1 A0A3Q2HLU3 A0A2U4C119 A0A2K6H024 A0A3Q2IDU3 A0A2U4C0Y1 F6WF49 A0A2Y9S8T1 L8IWX3 A0A2Y9S7Q2 A0A2Y9LTS1 A0A2U3V074 A0A2U4C0S9 A0A384ACM9 A0A2Y9LQT8 G5E630 A0A384ACG5 A0A452FL98 W5NVJ3 F7CE06 A0A146KRP5 A0A146LHV0 A0A2U4C0W6 A0A2Y9LYK2 G3TXD1 A0A2Y9S0S0 G3T223 U3JFP5 A0A287AC95 A0A287AM69 E9PWA9 Q8C7T5 A0A287A891 A0A287B4Y6 V4AR29 G3GSH2 A0A0N8CJH9 I3LMY9 A0A0P5ZXL4 A0A0P5U0S7 A0A0P6GU97 A0A0P5CYN1 A0A0P5AML0 A0A0P5VQM3 A0A0N8CJW0 A0A3L7H1V6
Pubmed
19121390
22118469
26354079
24845553
26334808
30249741
+ More
20798317 28648823 20075255 23537049 18362917 19820115 21719571 21347285 28004739 18563158 28503490 20566863 19892987 22751099 19393038 20809919 26823975 19468303 21183079 10349636 11042159 11076861 11217851 12466851 16141073 23254933 21804562 30723633 29704459
20798317 28648823 20075255 23537049 18362917 19820115 21719571 21347285 28004739 18563158 28503490 20566863 19892987 22751099 19393038 20809919 26823975 19468303 21183079 10349636 11042159 11076861 11217851 12466851 16141073 23254933 21804562 30723633 29704459
EMBL
BABH01030153
BABH01030154
BABH01030155
AGBW02013542
OWR43235.1
NWSH01004559
+ More
PCG64934.1 PCG64933.1 RSAL01000033 RVE51433.1 ODYU01011426 SOQ57158.1 KQ459602 KPI93409.1 KQ460615 KPJ13664.1 KQ760706 OAD59322.1 KZ288305 PBC28777.1 GFTR01008974 JAW07452.1 GECZ01026966 JAS42803.1 KK852413 KDR24425.1 GBGD01000019 JAC88870.1 NEVH01008214 PNF34529.1 GECU01029977 JAS77729.1 PNF34528.1 LBMM01002916 KMQ94190.1 GEDC01027569 GEDC01006879 JAS09729.1 JAS30419.1 QOIP01000005 RLU22895.1 KQ434809 KZC06489.1 KQ414894 KOC59763.1 GL436519 EFN71868.1 NNAY01000700 OXU26914.1 KQ978317 KYM95273.1 KB632303 ERL91819.1 APGK01018912 KB740085 ENN81554.1 KQ971342 EFA03030.2 KYB27698.1 KQ978579 KYN30005.1 GL887813 EGI69955.1 KQ982708 KYQ51790.1 KQ976453 KYM85441.1 ADTU01017494 KQ981920 KYN33012.1 LJIG01022814 KRT78580.1 GEZM01044529 GEZM01044528 JAV78164.1 IACN01088329 LAB59675.1 GG666612 EEN48999.1 GFAC01000854 JAT98334.1 NCKV01001752 RWS27831.1 GGLE01003915 MBY08041.1 RQTK01001514 RUS70007.1 AK294828 BAG57940.1 KQ435736 KOX77091.1 DS235044 EEB10914.1 PPHD01000318 POI35627.1 JH880514 ELR60956.1 GAPO01000002 JAB84491.1 AMGL01114840 AMGL01114841 GDHC01020989 JAP97639.1 GDHC01011704 JAQ06925.1 AGTO01001101 AGTO01011221 CU856620 FO082392 FO082421 FP565925 FQ440020 FQ456870 FQ670174 FQ976616 FQ976646 AL669967 AK049283 BAC33657.2 KB201549 ESO96146.1 JH000011 EGV96558.1 GDIP01125615 JAL78099.1 DQIR01171567 HDB27044.1 GDIP01050578 JAM53137.1 GDIP01123722 JAL79992.1 GDIQ01029298 JAN65439.1 GDIP01165159 JAJ58243.1 GDIP01196808 JAJ26594.1 GDIP01111268 JAL92446.1 GDIP01124567 JAL79147.1 RAZU01000328 RLQ59908.1
PCG64934.1 PCG64933.1 RSAL01000033 RVE51433.1 ODYU01011426 SOQ57158.1 KQ459602 KPI93409.1 KQ460615 KPJ13664.1 KQ760706 OAD59322.1 KZ288305 PBC28777.1 GFTR01008974 JAW07452.1 GECZ01026966 JAS42803.1 KK852413 KDR24425.1 GBGD01000019 JAC88870.1 NEVH01008214 PNF34529.1 GECU01029977 JAS77729.1 PNF34528.1 LBMM01002916 KMQ94190.1 GEDC01027569 GEDC01006879 JAS09729.1 JAS30419.1 QOIP01000005 RLU22895.1 KQ434809 KZC06489.1 KQ414894 KOC59763.1 GL436519 EFN71868.1 NNAY01000700 OXU26914.1 KQ978317 KYM95273.1 KB632303 ERL91819.1 APGK01018912 KB740085 ENN81554.1 KQ971342 EFA03030.2 KYB27698.1 KQ978579 KYN30005.1 GL887813 EGI69955.1 KQ982708 KYQ51790.1 KQ976453 KYM85441.1 ADTU01017494 KQ981920 KYN33012.1 LJIG01022814 KRT78580.1 GEZM01044529 GEZM01044528 JAV78164.1 IACN01088329 LAB59675.1 GG666612 EEN48999.1 GFAC01000854 JAT98334.1 NCKV01001752 RWS27831.1 GGLE01003915 MBY08041.1 RQTK01001514 RUS70007.1 AK294828 BAG57940.1 KQ435736 KOX77091.1 DS235044 EEB10914.1 PPHD01000318 POI35627.1 JH880514 ELR60956.1 GAPO01000002 JAB84491.1 AMGL01114840 AMGL01114841 GDHC01020989 JAP97639.1 GDHC01011704 JAQ06925.1 AGTO01001101 AGTO01011221 CU856620 FO082392 FO082421 FP565925 FQ440020 FQ456870 FQ670174 FQ976616 FQ976646 AL669967 AK049283 BAC33657.2 KB201549 ESO96146.1 JH000011 EGV96558.1 GDIP01125615 JAL78099.1 DQIR01171567 HDB27044.1 GDIP01050578 JAM53137.1 GDIP01123722 JAL79992.1 GDIQ01029298 JAN65439.1 GDIP01165159 JAJ58243.1 GDIP01196808 JAJ26594.1 GDIP01111268 JAL92446.1 GDIP01124567 JAL79147.1 RAZU01000328 RLQ59908.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000242457 UP000005203 UP000027135 UP000235965 UP000036403 UP000279307 UP000076502 UP000053825 UP000000311 UP000215335 UP000002358 UP000078542 UP000030742 UP000019118 UP000007266 UP000078492 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000001554 UP000288716 UP000271974 UP000085678 UP000053105 UP000233160 UP000009046 UP000002281 UP000245320 UP000248484 UP000248483 UP000261681 UP000009136 UP000291000 UP000002356 UP000007646 UP000016665 UP000008227 UP000000589 UP000030746 UP000001075 UP000273346
UP000242457 UP000005203 UP000027135 UP000235965 UP000036403 UP000279307 UP000076502 UP000053825 UP000000311 UP000215335 UP000002358 UP000078542 UP000030742 UP000019118 UP000007266 UP000078492 UP000007755 UP000075809 UP000078540 UP000005205 UP000078541 UP000001554 UP000288716 UP000271974 UP000085678 UP000053105 UP000233160 UP000009046 UP000002281 UP000245320 UP000248484 UP000248483 UP000261681 UP000009136 UP000291000 UP000002356 UP000007646 UP000016665 UP000008227 UP000000589 UP000030746 UP000001075 UP000273346
Interpro
Gene 3D
ProteinModelPortal
H9JDB0
A0A212EP08
A0A2A4J087
A0A2A4J0N2
A0A3S2TP78
A0A2H1WVS5
+ More
A0A194PL04 A0A194R833 A0A310SNR3 A0A2A3EAM0 A0A088A5Z8 A0A224XFS3 A0A1B6EYR0 A0A067RUD8 A0A069DY94 A0A2J7R122 A0A1B6HSU7 A0A2J7R116 A0A0J7NNW5 A0A1B6C8S8 A0A3L8DR29 A0A154P3H2 A0A0L7QM89 E2A416 A0A232F7U1 K7IRB1 A0A195C536 U4UCY8 N6TM76 D6WKR1 A0A139WI67 A0A151JRZ4 F4W771 A0A151WV29 A0A195BJS5 A0A158NJ55 A0A195EYM8 A0A0T6AU90 A0A1Y1LX88 A0A2D4PNU0 C3ZE43 A0A1E1XGF4 A0A443SJY2 A0A2R5LEU6 A0A433SLI9 B4DGX9 A0A2R2MQM1 A0A0N0BI91 A0A1S3IUS1 A0A2K6GZY0 E0VC08 A0A2P4TGZ1 A0A3Q2HLU3 A0A2U4C119 A0A2K6H024 A0A3Q2IDU3 A0A2U4C0Y1 F6WF49 A0A2Y9S8T1 L8IWX3 A0A2Y9S7Q2 A0A2Y9LTS1 A0A2U3V074 A0A2U4C0S9 A0A384ACM9 A0A2Y9LQT8 G5E630 A0A384ACG5 A0A452FL98 W5NVJ3 F7CE06 A0A146KRP5 A0A146LHV0 A0A2U4C0W6 A0A2Y9LYK2 G3TXD1 A0A2Y9S0S0 G3T223 U3JFP5 A0A287AC95 A0A287AM69 E9PWA9 Q8C7T5 A0A287A891 A0A287B4Y6 V4AR29 G3GSH2 A0A0N8CJH9 I3LMY9 A0A0P5ZXL4 A0A0P5U0S7 A0A0P6GU97 A0A0P5CYN1 A0A0P5AML0 A0A0P5VQM3 A0A0N8CJW0 A0A3L7H1V6
A0A194PL04 A0A194R833 A0A310SNR3 A0A2A3EAM0 A0A088A5Z8 A0A224XFS3 A0A1B6EYR0 A0A067RUD8 A0A069DY94 A0A2J7R122 A0A1B6HSU7 A0A2J7R116 A0A0J7NNW5 A0A1B6C8S8 A0A3L8DR29 A0A154P3H2 A0A0L7QM89 E2A416 A0A232F7U1 K7IRB1 A0A195C536 U4UCY8 N6TM76 D6WKR1 A0A139WI67 A0A151JRZ4 F4W771 A0A151WV29 A0A195BJS5 A0A158NJ55 A0A195EYM8 A0A0T6AU90 A0A1Y1LX88 A0A2D4PNU0 C3ZE43 A0A1E1XGF4 A0A443SJY2 A0A2R5LEU6 A0A433SLI9 B4DGX9 A0A2R2MQM1 A0A0N0BI91 A0A1S3IUS1 A0A2K6GZY0 E0VC08 A0A2P4TGZ1 A0A3Q2HLU3 A0A2U4C119 A0A2K6H024 A0A3Q2IDU3 A0A2U4C0Y1 F6WF49 A0A2Y9S8T1 L8IWX3 A0A2Y9S7Q2 A0A2Y9LTS1 A0A2U3V074 A0A2U4C0S9 A0A384ACM9 A0A2Y9LQT8 G5E630 A0A384ACG5 A0A452FL98 W5NVJ3 F7CE06 A0A146KRP5 A0A146LHV0 A0A2U4C0W6 A0A2Y9LYK2 G3TXD1 A0A2Y9S0S0 G3T223 U3JFP5 A0A287AC95 A0A287AM69 E9PWA9 Q8C7T5 A0A287A891 A0A287B4Y6 V4AR29 G3GSH2 A0A0N8CJH9 I3LMY9 A0A0P5ZXL4 A0A0P5U0S7 A0A0P6GU97 A0A0P5CYN1 A0A0P5AML0 A0A0P5VQM3 A0A0N8CJW0 A0A3L7H1V6
Ontologies
GO
Topology
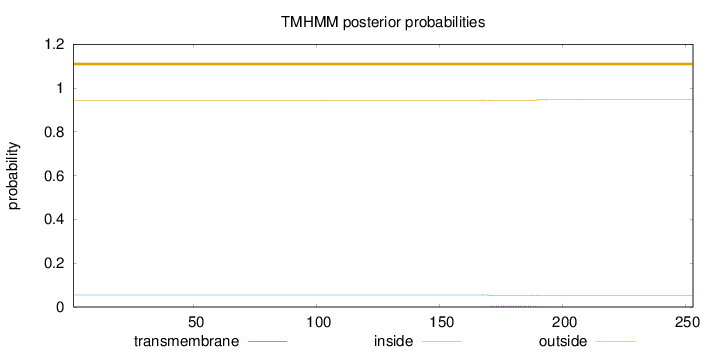
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09821
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05527
outside
1 - 253
Population Genetic Test Statistics
Pi
329.153584
Theta
196.362636
Tajima's D
1.869698
CLR
0
CSRT
0.860206989650517
Interpretation
Uncertain
