Gene
KWMTBOMO09055 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007502
Annotation
PREDICTED:_NAD(P)_transhydrogenase?_mitochondrial-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.044
Sequence
CDS
ATGTGCGGATCCTCAAGAATGCTTCGTGCATATAAACAATTACCAAGATCATGGCAATGCAGACTGTCAAGTAGCTCTTCAGCTACGCCTCCGTCAACCTCAGGAGTTTCTTACACCAAACTAAGCATAGGAGTGCCAAAAGAGATTTGGCAAGATGAAAGAAGGGTTGCAGTAGTACCAGCCGTAGTCAGTAAATTAGTAAAAAAAGGCTTTAATGTCAATATTGAAGAAGATGCAGGTGCTGTAGCTAATTTTCCTAATAAGACTTATGAAGAGGCTGGAGCAAAAATTACAAACTTAAAAACTACATTCCAATCTGATATAGTTTTGAAAGTTCGTCCACTCCTAAATAATGAAATACAAAATGTAAGGAATGAAGGAACATTGATATCTTTCCTCTATCCTGCTCAAAATCAAGATTTAATAAAAAAGCTTTCTGAAAGAAAGATGAATGCATTTGCTATGGACTGCATTCCCCGTATAAGTCGCGCACAGGCATTTGATGCTCTTAGTTCAATGGCAAATGTTGCTGGTTATCGTGCAGTTATTGAAGCTGCCGCTCATTTCCCACGTTTCTTTTCCGGACAAATGACGGCGGCTGGTCGTGTGCCTCCATGTCGTGTGCTGGTAGTGGGCGGGGGCGTGGCCGGACTGGCGGCCGCAGCGCAGGCCCGGTGCATGGGGGCCGCCGTGCGGGCCTTCGACACGCGTCCCGCCGTTCGTGAGCAAATCGAAAGTCTCGGAGCCCAATTCATTACCATGGAAATGAAGGAAGAGGGAGCCGGCGAAGGTGGGTACGCAAAAGAGATGAGTGAAGACTTCCTAAATGCTGAACGAGCTTTGCTGGGAAAAGAAACGAGATCTTCTGACGTTGTGATAAGTACAGCGCTTATACCTGGGAAACCTGCACCTTTACTAATTTTAGAGGACGCGGTTCGAGACATGGCACCTGGTAGCGTCATAGTAGACTTGGCAGCTGAAATGGGGGGCAACATTCAGACCACGAAGAAAGGGGAGATCACTAAAGTTCACGGCGTCACTCATATCGGCCTCACCGACCTGCCGAGCCGCATGCCCGCCCACGCTTCTACGTTGTATGCTAATAATATATCAGGATTTCTGTTGAGCTTGGGTACCAATGATCATTTTGATATCAACCTGGAGGACGAAGTGACCCGCGGCGCGATTGTCCTGAGAGCTGGCGAGCTGCTGTGGCCGCCCCCGCCGCCGCCCAGCATGGCGCCCGCACACGCCGCGCCCAAACCGACAGCCGTACAACCTGAACCACCGAACCCATTTAAAGACACTCTGAAGGACGCCTTTTTGTACTCAACAGGTATGGCGTCGTTGATCGGTTTGGGTATAGCTGCGCCGAATCCGGCGTTCACAACAATGACCACTACGCTAGCGTTGTCAGGCGTGGTCGGGTACCACACTGTGTGGGGCGTCGTGCCAGCTCTTCACTCGCCGCTCATGTCCGTTACAAACGCCGTGTCGGGTATTACGGCCGTCGGTGGACTACTCCTAATGGGCGGAGGATATGTTCCGGAGACGCCCGTTCAGTGGTTGGCGGGTACTGCAGCTTTAATATCTTTCGTCAATGTATTTGGAGGGTTCTTGGTTACGCAACGGATGCTTGATATGTTCAAGAGACCTGGTGATCCACCGGAATATGGTTATCTCTATGCAATTCCTGCCGCTGCGCTGCTCGGCGGCTACATCACTACCGCAATGCAGGGATATCCAGAGGTACACCAAATGGCTTATCTTGCTTCATCTTTGTGTTGTGTTGGAGCTCTTGCGGGACTCAGTTCTCAGACAACGGCACGAAAAGGAAACTATTTGGGAATAATCGGTGTCACTGGTGGTATCGCAGCCACCCTTGGAGCCTTGACTCCAAGTGCTGAAGTGCTGGCGCAAATGATAGGCGTAGCTGGCATTGGAGGTTTAATTGGAAGCACTATTGCTAAGAAAATAGAGATTACAGACCTTCCCCAACTCGTGGCTGGTTTCCACAGTTTGGTAGGTATGGCGGCAGTCCTAACCTGCATCGCTACATACATGCACGACTTCCCCGCAATGGCCTTGGATCCTACGGCAGCCACGCTCAAGACTTCGTTGTTCCTTGGGACATACATCGGTGGCATAACATTCACGGGCTCGCTAGTAGCGTACGGCAAGCTGCAAGGCTCGTTGTCGTCGGCGCCGCTGCTGCTGCCGGGTCGCCACGCCATCAACGCGGCTCTGCTGACGGGCGCGCTGGGCTGCGGCGGGGCACTGCTCGCCTTCCCCGAGGCGCCCGGCCTGCCGCTGCTGTCCGCTGCTGCACTGCTCAGCGGGATACAGGGGCTCACGCTTACCGCCGCCATTGGGGGTGCGGACATGCCCGTAGTGATCACGGTATTGAATAGCTACTCAGGATGGGCGCTGTGCGCTGAAGGTTTTATGTTAAACAACTCGCTCATGACTATAGTTGGCGCGCTCATTGGTAGTTCTGGTGCTATCTTGTCGTACATAATGTGCAAAGCAATGAACAGATCTCTCCCCAACGTCATCTTGGGTGGTTATGGAGTGACGACAGGTGGATCAGCTCGACCCACCGACGCGACACACACAGAACTCAATGTAGACTCAGTAGCCGAGCTTATCCACCGCGCGTCAAATATCATTATAACACCAGGTTATGGACTTTGCGTTGCGAAAGCGCAATACCCTATCGCTGAGTTAGTGGATCTTCTAAAGGAAAGCGGCAAGAAAGTGCGTTTTGCTATTCACCCTGTTGCAGGTCGTATGCCCGGACAACTCAACGTATTATTGGCCGAAGCTGGAGTTCCATACGATGACGTATTTGAAATGGATGAAATTAATGATGAATTCCCCGAGACTGATTTAGTACTTGTTATCGGTGCTAATGACACTGTAAATAGCGCGGCCGAAGATGATCCGAACTCACCTATTGCGGGAATGCCGGTACTCAAAGTATGGAAAGCGAATCAGGTTGTAGTAATGAAAAGATCAATGGGAGTTGGATATGCGGCCGTGGACAACCCGATATTCTACAATCCCAACACTGCGATGTTACTCGGCGATGCTAAAAAGACTTGTGACTCTCTACTTGAACGAGTTAAACATTTAGCAGCCTAA
Protein
MCGSSRMLRAYKQLPRSWQCRLSSSSSATPPSTSGVSYTKLSIGVPKEIWQDERRVAVVPAVVSKLVKKGFNVNIEEDAGAVANFPNKTYEEAGAKITNLKTTFQSDIVLKVRPLLNNEIQNVRNEGTLISFLYPAQNQDLIKKLSERKMNAFAMDCIPRISRAQAFDALSSMANVAGYRAVIEAAAHFPRFFSGQMTAAGRVPPCRVLVVGGGVAGLAAAAQARCMGAAVRAFDTRPAVREQIESLGAQFITMEMKEEGAGEGGYAKEMSEDFLNAERALLGKETRSSDVVISTALIPGKPAPLLILEDAVRDMAPGSVIVDLAAEMGGNIQTTKKGEITKVHGVTHIGLTDLPSRMPAHASTLYANNISGFLLSLGTNDHFDINLEDEVTRGAIVLRAGELLWPPPPPPSMAPAHAAPKPTAVQPEPPNPFKDTLKDAFLYSTGMASLIGLGIAAPNPAFTTMTTTLALSGVVGYHTVWGVVPALHSPLMSVTNAVSGITAVGGLLLMGGGYVPETPVQWLAGTAALISFVNVFGGFLVTQRMLDMFKRPGDPPEYGYLYAIPAAALLGGYITTAMQGYPEVHQMAYLASSLCCVGALAGLSSQTTARKGNYLGIIGVTGGIAATLGALTPSAEVLAQMIGVAGIGGLIGSTIAKKIEITDLPQLVAGFHSLVGMAAVLTCIATYMHDFPAMALDPTAATLKTSLFLGTYIGGITFTGSLVAYGKLQGSLSSAPLLLPGRHAINAALLTGALGCGGALLAFPEAPGLPLLSAAALLSGIQGLTLTAAIGGADMPVVITVLNSYSGWALCAEGFMLNNSLMTIVGALIGSSGAILSYIMCKAMNRSLPNVILGGYGVTTGGSARPTDATHTELNVDSVAELIHRASNIIITPGYGLCVAKAQYPIAELVDLLKESGKKVRFAIHPVAGRMPGQLNVLLAEAGVPYDDVFEMDEINDEFPETDLVLVIGANDTVNSAAEDDPNSPIAGMPVLKVWKANQVVVMKRSMGVGYAAVDNPIFYNPNTAMLLGDAKKTCDSLLERVKHLAA
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9JDA5
A0A3S2PG94
A0A2A4J724
A0A2H1WMG8
A0A194PJ99
A0A212EP35
+ More
A0A194R7N3 A0A182IT48 A0A336M0T5 U5ESM1 A0A1B0CS25 A0A1L8DZU8 A0A182MKN5 A0A1L8E0C8 D6WMM8 A0A2M4A3C1 A0A182QY46 A0A182FQB8 A0A2J7QTH8 W5JVG0 A0A182VXG6 A0A182NFS2 A0A1B6FCA9 A0A182SYL1 A0A182R386 A0A1Y1KCD6 A0A182PAH5 A0A182K4I1 A0A182GWF2 A0A182V0K9 A0A1S4FX45 Q16LL0 A0A182XE68 A0A182UEA2 Q7QBH8 A0A182HJ55 A0A0N7Z9U0 A0A067QXG1 A0A1Q3FIE5 A0A1D2MSL9 A0A286RXT6 A0A164X3C5 A0A0P6ICA7 B0W8J7 E0VR86 A0A0P6AY07 A0A182XWT9 E9GAE5 A0A1J1J5S9 A0A3R7QM12 A0A2R5LKY8 A0A3S3Q942 A0A146L8G8 A0A087UCJ2 A0A2T7PFS5 A0A023GMB2 A0A0P5TB14 T1JNF6 A0A131XKL7 L7LSB9 A0A2S2R5X4 A0A224YWD0 A0A0P5U116 A0A131YGD1 A0A0A9ZAR2 A0A1E1XFN7 A0A069DZP4 A0A131ZUV9 A0A023F448 A0A1X7V4Y0 A0A0N8AUY5 A0A0P5ILR7 A0A443SI29 A0A131XSI9 A0A0P5PI66 A0A0P5PJN6 V5HVJ9 J9JS99 A0A0N8C5D6 A0A0P5LJB9 A0A224XIV4 A0A0P5HYR1 A0A0P5HRC6 K1QU53 A0A0V0ZPH2 A0A0P5NK74 A0A0P5PFL5 A0A0V1L3M8 A0A0P5ANF9 A0A0P5HDP8 A0A0V0SBL4 A0A0K2SZ35 A0A0V1D863 A0A0V1NR46 A0A0V1L310 A0A0V0TWX0 W4YRG7 A0A0V0VG15 A0A0V1B3C7 A0A0V0SBJ0
A0A194R7N3 A0A182IT48 A0A336M0T5 U5ESM1 A0A1B0CS25 A0A1L8DZU8 A0A182MKN5 A0A1L8E0C8 D6WMM8 A0A2M4A3C1 A0A182QY46 A0A182FQB8 A0A2J7QTH8 W5JVG0 A0A182VXG6 A0A182NFS2 A0A1B6FCA9 A0A182SYL1 A0A182R386 A0A1Y1KCD6 A0A182PAH5 A0A182K4I1 A0A182GWF2 A0A182V0K9 A0A1S4FX45 Q16LL0 A0A182XE68 A0A182UEA2 Q7QBH8 A0A182HJ55 A0A0N7Z9U0 A0A067QXG1 A0A1Q3FIE5 A0A1D2MSL9 A0A286RXT6 A0A164X3C5 A0A0P6ICA7 B0W8J7 E0VR86 A0A0P6AY07 A0A182XWT9 E9GAE5 A0A1J1J5S9 A0A3R7QM12 A0A2R5LKY8 A0A3S3Q942 A0A146L8G8 A0A087UCJ2 A0A2T7PFS5 A0A023GMB2 A0A0P5TB14 T1JNF6 A0A131XKL7 L7LSB9 A0A2S2R5X4 A0A224YWD0 A0A0P5U116 A0A131YGD1 A0A0A9ZAR2 A0A1E1XFN7 A0A069DZP4 A0A131ZUV9 A0A023F448 A0A1X7V4Y0 A0A0N8AUY5 A0A0P5ILR7 A0A443SI29 A0A131XSI9 A0A0P5PI66 A0A0P5PJN6 V5HVJ9 J9JS99 A0A0N8C5D6 A0A0P5LJB9 A0A224XIV4 A0A0P5HYR1 A0A0P5HRC6 K1QU53 A0A0V0ZPH2 A0A0P5NK74 A0A0P5PFL5 A0A0V1L3M8 A0A0P5ANF9 A0A0P5HDP8 A0A0V0SBL4 A0A0K2SZ35 A0A0V1D863 A0A0V1NR46 A0A0V1L310 A0A0V0TWX0 W4YRG7 A0A0V0VG15 A0A0V1B3C7 A0A0V0SBJ0
Pubmed
EMBL
BABH01030170
RSAL01000046
RVE50545.1
NWSH01002582
PCG67927.1
ODYU01009673
+ More
SOQ54265.1 KQ459602 KPI93402.1 AGBW02013542 OWR43244.1 KQ460615 KPJ13657.1 UFQT01000294 SSX22931.1 GANO01002287 JAB57584.1 AJWK01025585 AJWK01025586 GFDF01002140 JAV11944.1 AXCM01011788 GFDF01002139 JAV11945.1 KQ971343 EFA03278.1 GGFK01001817 MBW35138.1 AXCN02000243 NEVH01011194 PNF31874.1 ADMH02000324 ETN66970.1 GECZ01021924 GECZ01000798 JAS47845.1 JAS68971.1 GEZM01088882 JAV57850.1 JXUM01094144 JXUM01094145 KQ564106 KXJ72786.1 CH477902 EAT35199.1 AAAB01008879 EAA08455.4 APCN01002125 GDRN01110024 GDRN01110023 JAI56984.1 KK852901 KDR14080.1 GFDL01007711 JAV27334.1 LJIJ01000610 ODM95891.1 KY974269 ASW35069.1 LRGB01001019 KZS13829.1 GDIQ01006666 JAN88071.1 DS231859 EDS39047.1 DS235459 EEB15892.1 GDIP01022757 JAM80958.1 GL732537 EFX83224.1 CVRI01000073 CRL07751.1 QCYY01000697 ROT83361.1 GGLE01006036 MBY10162.1 NCKU01000299 RWS16102.1 GDHC01013956 GDHC01002615 JAQ04673.1 JAQ16014.1 KK119210 KFM75081.1 PZQS01000004 PVD32272.1 GBBM01000434 JAC34984.1 GDIP01130074 JAL73640.1 JH431984 GEFH01001891 JAP66690.1 GACK01010088 JAA54946.1 GGMS01015559 MBY84762.1 GFPF01010089 MAA21235.1 GDIP01123630 JAL80084.1 GEDV01010203 JAP78354.1 GBHO01001242 GBRD01012589 GBRD01012585 GBRD01005078 GBRD01005076 JAG42362.1 JAG53235.1 GFAC01001302 JAT97886.1 GBGD01000285 JAC88604.1 JXLN01002203 KPM02534.1 GBBI01002442 JAC16270.1 GDIQ01240379 JAK11346.1 GDIQ01236332 JAK15393.1 NCKV01002203 RWS27178.1 GEFM01006631 JAP69165.1 GDIQ01127964 JAL23762.1 GDIQ01127967 JAL23759.1 GANP01004186 JAB80282.1 ABLF02021445 GDIQ01113171 JAL38555.1 GDIQ01179592 JAK72133.1 GFTR01008455 JAW07971.1 GDIQ01221284 JAK30441.1 GDIQ01224077 JAK27648.1 JH817892 EKC40377.1 JYDQ01000122 KRY14188.1 GDIQ01145120 GDIQ01114415 JAL06606.1 GDIQ01129271 JAL22455.1 JYDW01000153 KRZ53842.1 GDIP01196760 JAJ26642.1 GDIQ01235184 JAK16541.1 JYDL01000020 KRX24092.1 HACA01001120 CDW18481.1 JYDI01000029 KRY57620.1 JYDM01000123 KRZ86243.1 KRZ53841.1 JYDJ01000118 KRX43473.1 AAGJ04026856 JYDN01000041 KRX62157.1 JYDH01000119 KRY31389.1 KRX24091.1
SOQ54265.1 KQ459602 KPI93402.1 AGBW02013542 OWR43244.1 KQ460615 KPJ13657.1 UFQT01000294 SSX22931.1 GANO01002287 JAB57584.1 AJWK01025585 AJWK01025586 GFDF01002140 JAV11944.1 AXCM01011788 GFDF01002139 JAV11945.1 KQ971343 EFA03278.1 GGFK01001817 MBW35138.1 AXCN02000243 NEVH01011194 PNF31874.1 ADMH02000324 ETN66970.1 GECZ01021924 GECZ01000798 JAS47845.1 JAS68971.1 GEZM01088882 JAV57850.1 JXUM01094144 JXUM01094145 KQ564106 KXJ72786.1 CH477902 EAT35199.1 AAAB01008879 EAA08455.4 APCN01002125 GDRN01110024 GDRN01110023 JAI56984.1 KK852901 KDR14080.1 GFDL01007711 JAV27334.1 LJIJ01000610 ODM95891.1 KY974269 ASW35069.1 LRGB01001019 KZS13829.1 GDIQ01006666 JAN88071.1 DS231859 EDS39047.1 DS235459 EEB15892.1 GDIP01022757 JAM80958.1 GL732537 EFX83224.1 CVRI01000073 CRL07751.1 QCYY01000697 ROT83361.1 GGLE01006036 MBY10162.1 NCKU01000299 RWS16102.1 GDHC01013956 GDHC01002615 JAQ04673.1 JAQ16014.1 KK119210 KFM75081.1 PZQS01000004 PVD32272.1 GBBM01000434 JAC34984.1 GDIP01130074 JAL73640.1 JH431984 GEFH01001891 JAP66690.1 GACK01010088 JAA54946.1 GGMS01015559 MBY84762.1 GFPF01010089 MAA21235.1 GDIP01123630 JAL80084.1 GEDV01010203 JAP78354.1 GBHO01001242 GBRD01012589 GBRD01012585 GBRD01005078 GBRD01005076 JAG42362.1 JAG53235.1 GFAC01001302 JAT97886.1 GBGD01000285 JAC88604.1 JXLN01002203 KPM02534.1 GBBI01002442 JAC16270.1 GDIQ01240379 JAK11346.1 GDIQ01236332 JAK15393.1 NCKV01002203 RWS27178.1 GEFM01006631 JAP69165.1 GDIQ01127964 JAL23762.1 GDIQ01127967 JAL23759.1 GANP01004186 JAB80282.1 ABLF02021445 GDIQ01113171 JAL38555.1 GDIQ01179592 JAK72133.1 GFTR01008455 JAW07971.1 GDIQ01221284 JAK30441.1 GDIQ01224077 JAK27648.1 JH817892 EKC40377.1 JYDQ01000122 KRY14188.1 GDIQ01145120 GDIQ01114415 JAL06606.1 GDIQ01129271 JAL22455.1 JYDW01000153 KRZ53842.1 GDIP01196760 JAJ26642.1 GDIQ01235184 JAK16541.1 JYDL01000020 KRX24092.1 HACA01001120 CDW18481.1 JYDI01000029 KRY57620.1 JYDM01000123 KRZ86243.1 KRZ53841.1 JYDJ01000118 KRX43473.1 AAGJ04026856 JYDN01000041 KRX62157.1 JYDH01000119 KRY31389.1 KRX24091.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000075880 UP000092461 UP000075883 UP000007266 UP000075886 UP000069272 UP000235965 UP000000673 UP000075920 UP000075884 UP000075901 UP000075900 UP000075885 UP000075881 UP000069940 UP000249989 UP000075903 UP000008820 UP000076407 UP000075902 UP000007062 UP000075840 UP000027135 UP000094527 UP000076858 UP000002320 UP000009046 UP000076408 UP000000305 UP000183832 UP000283509 UP000285301 UP000054359 UP000245119 UP000007879 UP000288716 UP000007819 UP000005408 UP000054783 UP000054721 UP000054630 UP000054653 UP000054924 UP000055048 UP000007110 UP000054681 UP000054776
UP000075880 UP000092461 UP000075883 UP000007266 UP000075886 UP000069272 UP000235965 UP000000673 UP000075920 UP000075884 UP000075901 UP000075900 UP000075885 UP000075881 UP000069940 UP000249989 UP000075903 UP000008820 UP000076407 UP000075902 UP000007062 UP000075840 UP000027135 UP000094527 UP000076858 UP000002320 UP000009046 UP000076408 UP000000305 UP000183832 UP000283509 UP000285301 UP000054359 UP000245119 UP000007879 UP000288716 UP000007819 UP000005408 UP000054783 UP000054721 UP000054630 UP000054653 UP000054924 UP000055048 UP000007110 UP000054681 UP000054776
PRIDE
Pfam
Interpro
IPR034300
PNTB-like
+ More
IPR026255 NADP_transhyd_a
IPR007698 AlaDH/PNT_NAD(H)-bd
IPR036291 NAD(P)-bd_dom_sf
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR007886 AlaDH/PNT_N
IPR024605 NADP_transhyd_a_C
IPR039010 Synaptotagmin_SMP
IPR031468 SMP_LBD
IPR000008 C2_dom
IPR037749 Ext_Synaptotagmin_C2B
IPR035892 C2_domain_sf
IPR037752 C2C_KIAA1228
IPR008143 Ala_DH/PNT_CS2
IPR008142 AlaDH/PNT_CS1
IPR031734 MBF2
IPR036396 Cyt_P450_sf
IPR002402 Cyt_P450_E_grp-II
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR002300 aa-tRNA-synth_Ia
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
IPR002302 Leu-tRNA-ligase
IPR026255 NADP_transhyd_a
IPR007698 AlaDH/PNT_NAD(H)-bd
IPR036291 NAD(P)-bd_dom_sf
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR007886 AlaDH/PNT_N
IPR024605 NADP_transhyd_a_C
IPR039010 Synaptotagmin_SMP
IPR031468 SMP_LBD
IPR000008 C2_dom
IPR037749 Ext_Synaptotagmin_C2B
IPR035892 C2_domain_sf
IPR037752 C2C_KIAA1228
IPR008143 Ala_DH/PNT_CS2
IPR008142 AlaDH/PNT_CS1
IPR031734 MBF2
IPR036396 Cyt_P450_sf
IPR002402 Cyt_P450_E_grp-II
IPR001128 Cyt_P450
IPR017972 Cyt_P450_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR002300 aa-tRNA-synth_Ia
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
IPR002302 Leu-tRNA-ligase
Gene 3D
ProteinModelPortal
H9JDA5
A0A3S2PG94
A0A2A4J724
A0A2H1WMG8
A0A194PJ99
A0A212EP35
+ More
A0A194R7N3 A0A182IT48 A0A336M0T5 U5ESM1 A0A1B0CS25 A0A1L8DZU8 A0A182MKN5 A0A1L8E0C8 D6WMM8 A0A2M4A3C1 A0A182QY46 A0A182FQB8 A0A2J7QTH8 W5JVG0 A0A182VXG6 A0A182NFS2 A0A1B6FCA9 A0A182SYL1 A0A182R386 A0A1Y1KCD6 A0A182PAH5 A0A182K4I1 A0A182GWF2 A0A182V0K9 A0A1S4FX45 Q16LL0 A0A182XE68 A0A182UEA2 Q7QBH8 A0A182HJ55 A0A0N7Z9U0 A0A067QXG1 A0A1Q3FIE5 A0A1D2MSL9 A0A286RXT6 A0A164X3C5 A0A0P6ICA7 B0W8J7 E0VR86 A0A0P6AY07 A0A182XWT9 E9GAE5 A0A1J1J5S9 A0A3R7QM12 A0A2R5LKY8 A0A3S3Q942 A0A146L8G8 A0A087UCJ2 A0A2T7PFS5 A0A023GMB2 A0A0P5TB14 T1JNF6 A0A131XKL7 L7LSB9 A0A2S2R5X4 A0A224YWD0 A0A0P5U116 A0A131YGD1 A0A0A9ZAR2 A0A1E1XFN7 A0A069DZP4 A0A131ZUV9 A0A023F448 A0A1X7V4Y0 A0A0N8AUY5 A0A0P5ILR7 A0A443SI29 A0A131XSI9 A0A0P5PI66 A0A0P5PJN6 V5HVJ9 J9JS99 A0A0N8C5D6 A0A0P5LJB9 A0A224XIV4 A0A0P5HYR1 A0A0P5HRC6 K1QU53 A0A0V0ZPH2 A0A0P5NK74 A0A0P5PFL5 A0A0V1L3M8 A0A0P5ANF9 A0A0P5HDP8 A0A0V0SBL4 A0A0K2SZ35 A0A0V1D863 A0A0V1NR46 A0A0V1L310 A0A0V0TWX0 W4YRG7 A0A0V0VG15 A0A0V1B3C7 A0A0V0SBJ0
A0A194R7N3 A0A182IT48 A0A336M0T5 U5ESM1 A0A1B0CS25 A0A1L8DZU8 A0A182MKN5 A0A1L8E0C8 D6WMM8 A0A2M4A3C1 A0A182QY46 A0A182FQB8 A0A2J7QTH8 W5JVG0 A0A182VXG6 A0A182NFS2 A0A1B6FCA9 A0A182SYL1 A0A182R386 A0A1Y1KCD6 A0A182PAH5 A0A182K4I1 A0A182GWF2 A0A182V0K9 A0A1S4FX45 Q16LL0 A0A182XE68 A0A182UEA2 Q7QBH8 A0A182HJ55 A0A0N7Z9U0 A0A067QXG1 A0A1Q3FIE5 A0A1D2MSL9 A0A286RXT6 A0A164X3C5 A0A0P6ICA7 B0W8J7 E0VR86 A0A0P6AY07 A0A182XWT9 E9GAE5 A0A1J1J5S9 A0A3R7QM12 A0A2R5LKY8 A0A3S3Q942 A0A146L8G8 A0A087UCJ2 A0A2T7PFS5 A0A023GMB2 A0A0P5TB14 T1JNF6 A0A131XKL7 L7LSB9 A0A2S2R5X4 A0A224YWD0 A0A0P5U116 A0A131YGD1 A0A0A9ZAR2 A0A1E1XFN7 A0A069DZP4 A0A131ZUV9 A0A023F448 A0A1X7V4Y0 A0A0N8AUY5 A0A0P5ILR7 A0A443SI29 A0A131XSI9 A0A0P5PI66 A0A0P5PJN6 V5HVJ9 J9JS99 A0A0N8C5D6 A0A0P5LJB9 A0A224XIV4 A0A0P5HYR1 A0A0P5HRC6 K1QU53 A0A0V0ZPH2 A0A0P5NK74 A0A0P5PFL5 A0A0V1L3M8 A0A0P5ANF9 A0A0P5HDP8 A0A0V0SBL4 A0A0K2SZ35 A0A0V1D863 A0A0V1NR46 A0A0V1L310 A0A0V0TWX0 W4YRG7 A0A0V0VG15 A0A0V1B3C7 A0A0V0SBJ0
PDB
1U31
E-value=8.45757e-85,
Score=803
Ontologies
PATHWAY
GO
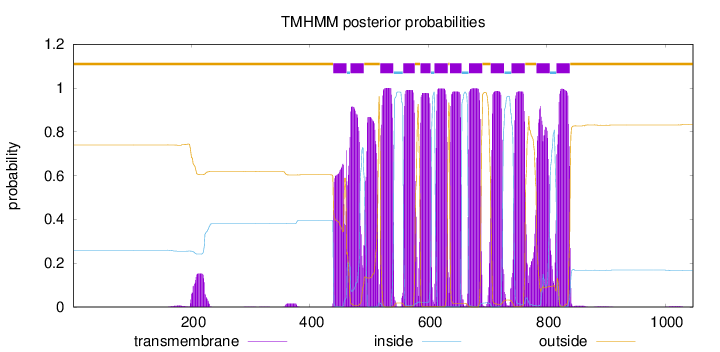
Topology
Length:
1047
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
265.66166
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.25968
outside
1 - 439
TMhelix
440 - 462
inside
463 - 468
TMhelix
469 - 491
outside
492 - 518
TMhelix
519 - 541
inside
542 - 557
TMhelix
558 - 577
outside
578 - 586
TMhelix
587 - 604
inside
605 - 610
TMhelix
611 - 633
outside
634 - 636
TMhelix
637 - 656
inside
657 - 668
TMhelix
669 - 691
outside
692 - 705
TMhelix
706 - 728
inside
729 - 740
TMhelix
741 - 763
outside
764 - 782
TMhelix
783 - 805
inside
806 - 816
TMhelix
817 - 839
outside
840 - 1047
Population Genetic Test Statistics
Pi
243.800063
Theta
177.779991
Tajima's D
1.14184
CLR
0.015041
CSRT
0.696315184240788
Interpretation
Uncertain
