Gene
KWMTBOMO09054 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007586
Annotation
NADPH-specific_isocitrate_dehydrogenase_[Bombyx_mori]
Full name
Isocitrate dehydrogenase [NADP]
Location in the cell
Cytoplasmic Reliability : 3.431
Sequence
CDS
ATGGCTAATACTAGTACAACAAAAATTCTAAAGTGTGTAAACCAGTTTGCATCACTGACTAGAAACTATGGAACTGCTAAGAGAGTTGTGGCTGCTAAACCAGTTGTCGAGATGGATGGTGATGAGATGACACGCATCATCTGGGCTAAGATCAAAGAGAGGCTGATATTTCCATATGTTAAGGTGGATTGTCTTTACTTTGACTTGGGCCTCCCGCATCGTGACGCCACTGATGATCAGGTCACCATCGACTCAGCGCATGCCATTCTAAAGCATAATGTGGGCATCAAATGCGCTACTATCACACCTGATGAACAAAGAGTAGAAGAGTTCAAGTTGAAGAAAATGTGGCTAAGTCCGAACGGAACGATCCGTAACATTTTGGGCGGTACAGTATTCCGTGAACCGATATTGTGTCAGAGTATCCCGAGGGTTGTGCCTGGTTGGACGAAACCTATTGTGATAGGCCGTCATGCGCACGGCGACCAATACAAGGCGCAAGATTTTGTTGTACCGAAACCCGGAAAGGTTGAACTCGTTTACACTACACAAGATGGTACGACTGAGAGGCGTGTATTATATGATTTTAAAACTCCAGGCGTTGCTATGGGCATGTACAATACCGACGAATCGATTCGGTCCTTTGCCCATTCCAGTTTTCAGGTAGCTTTACAGAAAAAATGGCCTCTATACCTGTCCACGAAGAATACCATTTTGAAACGTTACGATGGTCGTTTCAAAGACATTTTCGAAGAAGTTTTTCAGAGTGATTACAAGACTAAGTTTGATGAAGCCAAGATATGGTACGAGCATCGTTTAATTGACGACATGGTGGCACAGGCTATTAAGGGATCAGGTGGCTTTGTTTGGGCCTGCAAAAACTACGACGGAGATGTGCAGTCCGATATTGTTGCTCAGGGTTACGGGTCATTGGGAATGATGACATCAGTATTGATGTGTCCCGATGGCCGTACCGTGGAATCCGAAGCGGCGCACGGGACGGTGACGCGTCACTACCGTATGCACCAGCAAGGCAAGCCGACATCGACTAACCCGGTCGCTTCTATCTACGCCTGGACCAGAGGCCTTGCACACCGGGCCAAATTAGACGGGACTCCCGAATTGGAGCGCTTCGCTCTAGCCCTCGAAGAGGCGTGCGTCGAGTGCATCGATAGCGGTAAAATGACAAAAGATTTGGTTATTTGTATACACGGATTGGCGAACACGAAAGAAGGTATGTTTTTACACACCGAAGATTTCCTGGAGGCCATCGCCGAGCAGCTGGAACGCAAGTTGGGCGGCAAATAA
Protein
MANTSTTKILKCVNQFASLTRNYGTAKRVVAAKPVVEMDGDEMTRIIWAKIKERLIFPYVKVDCLYFDLGLPHRDATDDQVTIDSAHAILKHNVGIKCATITPDEQRVEEFKLKKMWLSPNGTIRNILGGTVFREPILCQSIPRVVPGWTKPIVIGRHAHGDQYKAQDFVVPKPGKVELVYTTQDGTTERRVLYDFKTPGVAMGMYNTDESIRSFAHSSFQVALQKKWPLYLSTKNTILKRYDGRFKDIFEEVFQSDYKTKFDEAKIWYEHRLIDDMVAQAIKGSGGFVWACKNYDGDVQSDIVAQGYGSLGMMTSVLMCPDGRTVESEAAHGTVTRHYRMHQQGKPTSTNPVASIYAWTRGLAHRAKLDGTPELERFALALEEACVECIDSGKMTKDLVICIHGLANTKEGMFLHTEDFLEAIAEQLERKLGGK
Summary
Catalytic Activity
isocitrate + NADP(+) = 2-oxoglutarate + CO2 + NADPH
Similarity
Belongs to the isocitrate and isopropylmalate dehydrogenases family.
Uniprot
Q1HQ47
Q2F682
S4PG03
C5IU79
A0A437BJH0
A0A194PJ03
+ More
A0A2A4J839 A0A2H1WMR5 A0A0L7KS43 A0A336L332 A0A1L8E0T4 D6WMM9 U5EUN0 A0A2Z5RE44 A0A2J7QTH1 A0A182FQB7 A0A194RCL7 A0A182GWF1 A0A1Q3G3V0 W5JVS7 A0A0P6IWM1 A0A2M4AET7 A0A1B6DUK8 T1DNG1 A0A2M3ZH85 A0A2M4BN00 A0A182IQW4 A0A182MDB5 A0A182R385 A0A182TNU5 A0A182VLM9 A0A182XE67 A0A182LCC4 Q7PQQ3 A0A182HJ56 A0A067R707 A0A084VT63 A0A182PAH4 A0A182K1D2 A0A212EP23 A0A1B6H9E0 A0A182VXG5 A0A170ZT89 A0A182XWU0 A0A182QLM4 A0A1B0GPW4 A0A1B6L4T4 B0W8J6 R4WRJ5 A0A1Q3FH56 A0A1J1J8G1 A0A023ERF4 A0A182NFS3 A0A0P6GDX1 A0A164X3A6 T1HWW5 A0A0P5M0M1 V5G6Y6 A0A0P6IAQ5 A0A2S2P120 A0A0N7Z8U3 J9K5R3 A0A0C5Q4W6 E9GAE4 A0A069DU83 A0A3R7SZS6 A0A0P6CFA3 A0A023F9T3 A0A2P1JJ64 A0A224XDB6 A0A2H8TLD4 A0A1Y1KBI4 A0A0L8H5J4 R4G2N3 A0A0V0GA37 A0A182SBQ9 D3PJB8 A0A0P5G6F5 A0A1D2MS27 A0A226ELI4 A0A2L2XYP2 A0A0P5ZHN6 U4UK34 J3JWC0 V3ZNV5 A0A0N8CY74 E0VCW6 N6U0Y1 A0A1I8JHX3 A0A087TI66 A0A1I8IEN0 T1JA51 A0A1I8IA73 A0A2T7PFS0 A0A0V1DFM4 A0A267E1V2 K1RZE2 A0A0V1DGP9 A0A023GKA1 I1VYX6
A0A2A4J839 A0A2H1WMR5 A0A0L7KS43 A0A336L332 A0A1L8E0T4 D6WMM9 U5EUN0 A0A2Z5RE44 A0A2J7QTH1 A0A182FQB7 A0A194RCL7 A0A182GWF1 A0A1Q3G3V0 W5JVS7 A0A0P6IWM1 A0A2M4AET7 A0A1B6DUK8 T1DNG1 A0A2M3ZH85 A0A2M4BN00 A0A182IQW4 A0A182MDB5 A0A182R385 A0A182TNU5 A0A182VLM9 A0A182XE67 A0A182LCC4 Q7PQQ3 A0A182HJ56 A0A067R707 A0A084VT63 A0A182PAH4 A0A182K1D2 A0A212EP23 A0A1B6H9E0 A0A182VXG5 A0A170ZT89 A0A182XWU0 A0A182QLM4 A0A1B0GPW4 A0A1B6L4T4 B0W8J6 R4WRJ5 A0A1Q3FH56 A0A1J1J8G1 A0A023ERF4 A0A182NFS3 A0A0P6GDX1 A0A164X3A6 T1HWW5 A0A0P5M0M1 V5G6Y6 A0A0P6IAQ5 A0A2S2P120 A0A0N7Z8U3 J9K5R3 A0A0C5Q4W6 E9GAE4 A0A069DU83 A0A3R7SZS6 A0A0P6CFA3 A0A023F9T3 A0A2P1JJ64 A0A224XDB6 A0A2H8TLD4 A0A1Y1KBI4 A0A0L8H5J4 R4G2N3 A0A0V0GA37 A0A182SBQ9 D3PJB8 A0A0P5G6F5 A0A1D2MS27 A0A226ELI4 A0A2L2XYP2 A0A0P5ZHN6 U4UK34 J3JWC0 V3ZNV5 A0A0N8CY74 E0VCW6 N6U0Y1 A0A1I8JHX3 A0A087TI66 A0A1I8IEN0 T1JA51 A0A1I8IA73 A0A2T7PFS0 A0A0V1DFM4 A0A267E1V2 K1RZE2 A0A0V1DGP9 A0A023GKA1 I1VYX6
EC Number
1.1.1.42
Pubmed
19121390
23622113
26354079
26227816
18362917
19820115
+ More
26760975 26483478 20920257 23761445 26999592 20966253 12364791 14747013 17210077 24845553 24438588 22118469 25244985 23691247 24945155 27129103 25702953 21292972 26334808 25474469 28004739 27289101 26561354 23537049 22516182 23254933 20566863 26392545 22992520 22491035
26760975 26483478 20920257 23761445 26999592 20966253 12364791 14747013 17210077 24845553 24438588 22118469 25244985 23691247 24945155 27129103 25702953 21292972 26334808 25474469 28004739 27289101 26561354 23537049 22516182 23254933 20566863 26392545 22992520 22491035
EMBL
BABH01030171
DQ443205
ABF51294.1
DQ311190
ABD36135.1
GAIX01003827
+ More
JAA88733.1 FJ959347 ACR46834.1 RSAL01000046 RVE50546.1 KQ459602 KPI93401.1 NWSH01002582 PCG67926.1 ODYU01009673 SOQ54266.1 JTDY01006605 KOB65844.1 UFQS01001784 UFQT01001784 SSX12346.1 SSX31797.1 GFDF01001773 JAV12311.1 KQ971343 EFA04299.1 GANO01002250 JAB57621.1 FX983736 BAX07235.1 NEVH01011194 PNF31876.1 KQ460615 KPJ13656.1 JXUM01094140 KQ564106 KXJ72785.1 GFDL01000563 JAV34482.1 ADMH02000324 ETN66969.1 GDUN01000615 JAN95304.1 GGFK01005827 MBW39148.1 GEDC01007945 JAS29353.1 GAMD01002915 JAA98675.1 GGFM01007176 MBW27927.1 GGFJ01005316 MBW54457.1 AXCM01007766 AAAB01008879 EAA08466.6 APCN01002125 KK852901 KDR14081.1 ATLV01016236 ATLV01016237 KE525072 KFB41157.1 AGBW02013542 OWR43245.1 GECU01036452 GECU01029594 GECU01008122 JAS71254.1 JAS78112.1 JAS99584.1 GEMB01001892 JAS01280.1 AXCN02000243 AJVK01064286 GEBQ01021312 JAT18665.1 DS231859 EDS39046.1 AK417312 BAN20527.1 GFDL01008159 JAV26886.1 CVRI01000073 CRL07750.1 GAPW01001833 JAC11765.1 GDIQ01034567 JAN60170.1 LRGB01001019 KZS13828.1 ACPB03018449 GDIQ01171422 JAK80303.1 GALX01002616 JAB65850.1 GDIQ01007229 JAN87508.1 GGMR01010423 MBY23042.1 GDKW01002387 JAI54208.1 ABLF02033188 KM609497 AJQ30121.1 GL732537 EFX83537.1 GBGD01001627 JAC87262.1 QCYY01000697 ROT83362.1 GDIP01016173 JAM87542.1 GBBI01000948 JAC17764.1 MF385062 AVN99057.1 GFTR01005991 JAW10435.1 GFXV01003149 MBW14954.1 GEZM01087167 JAV58873.1 KQ419114 KOF84561.1 GAHY01002456 JAA75054.1 GECL01001869 JAP04255.1 BT121724 HACA01002101 ADD38654.1 CDW19462.1 GDIQ01245752 JAK05973.1 LJIJ01000610 ODM95890.1 LNIX01000003 OXA58545.1 IAAA01001112 IAAA01001113 IAAA01001114 LAA01074.1 GDIP01043405 JAM60310.1 KB632357 ERL93482.1 BT127538 AEE62500.1 KB203188 ESO85992.1 GDIP01087255 JAM16460.1 DS235065 EEB11222.1 APGK01043801 KB741018 ENN75185.1 NIVC01001062 PAA72755.1 KK115333 KFM64805.1 JH431984 PZQS01000004 PVD32273.1 JYDI01000007 KRY60164.1 NIVC01002850 PAA54857.1 JH817892 EKC40376.1 KRY60163.1 GBBM01001107 JAC34311.1 JQ429383 AFI56368.1
JAA88733.1 FJ959347 ACR46834.1 RSAL01000046 RVE50546.1 KQ459602 KPI93401.1 NWSH01002582 PCG67926.1 ODYU01009673 SOQ54266.1 JTDY01006605 KOB65844.1 UFQS01001784 UFQT01001784 SSX12346.1 SSX31797.1 GFDF01001773 JAV12311.1 KQ971343 EFA04299.1 GANO01002250 JAB57621.1 FX983736 BAX07235.1 NEVH01011194 PNF31876.1 KQ460615 KPJ13656.1 JXUM01094140 KQ564106 KXJ72785.1 GFDL01000563 JAV34482.1 ADMH02000324 ETN66969.1 GDUN01000615 JAN95304.1 GGFK01005827 MBW39148.1 GEDC01007945 JAS29353.1 GAMD01002915 JAA98675.1 GGFM01007176 MBW27927.1 GGFJ01005316 MBW54457.1 AXCM01007766 AAAB01008879 EAA08466.6 APCN01002125 KK852901 KDR14081.1 ATLV01016236 ATLV01016237 KE525072 KFB41157.1 AGBW02013542 OWR43245.1 GECU01036452 GECU01029594 GECU01008122 JAS71254.1 JAS78112.1 JAS99584.1 GEMB01001892 JAS01280.1 AXCN02000243 AJVK01064286 GEBQ01021312 JAT18665.1 DS231859 EDS39046.1 AK417312 BAN20527.1 GFDL01008159 JAV26886.1 CVRI01000073 CRL07750.1 GAPW01001833 JAC11765.1 GDIQ01034567 JAN60170.1 LRGB01001019 KZS13828.1 ACPB03018449 GDIQ01171422 JAK80303.1 GALX01002616 JAB65850.1 GDIQ01007229 JAN87508.1 GGMR01010423 MBY23042.1 GDKW01002387 JAI54208.1 ABLF02033188 KM609497 AJQ30121.1 GL732537 EFX83537.1 GBGD01001627 JAC87262.1 QCYY01000697 ROT83362.1 GDIP01016173 JAM87542.1 GBBI01000948 JAC17764.1 MF385062 AVN99057.1 GFTR01005991 JAW10435.1 GFXV01003149 MBW14954.1 GEZM01087167 JAV58873.1 KQ419114 KOF84561.1 GAHY01002456 JAA75054.1 GECL01001869 JAP04255.1 BT121724 HACA01002101 ADD38654.1 CDW19462.1 GDIQ01245752 JAK05973.1 LJIJ01000610 ODM95890.1 LNIX01000003 OXA58545.1 IAAA01001112 IAAA01001113 IAAA01001114 LAA01074.1 GDIP01043405 JAM60310.1 KB632357 ERL93482.1 BT127538 AEE62500.1 KB203188 ESO85992.1 GDIP01087255 JAM16460.1 DS235065 EEB11222.1 APGK01043801 KB741018 ENN75185.1 NIVC01001062 PAA72755.1 KK115333 KFM64805.1 JH431984 PZQS01000004 PVD32273.1 JYDI01000007 KRY60164.1 NIVC01002850 PAA54857.1 JH817892 EKC40376.1 KRY60163.1 GBBM01001107 JAC34311.1 JQ429383 AFI56368.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000037510
UP000007266
+ More
UP000235965 UP000069272 UP000053240 UP000069940 UP000249989 UP000000673 UP000075880 UP000075883 UP000075900 UP000075902 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000027135 UP000030765 UP000075885 UP000075881 UP000007151 UP000075920 UP000076408 UP000075886 UP000092462 UP000002320 UP000183832 UP000075884 UP000076858 UP000015103 UP000007819 UP000000305 UP000283509 UP000053454 UP000075901 UP000094527 UP000198287 UP000030742 UP000030746 UP000009046 UP000019118 UP000095280 UP000215902 UP000054359 UP000245119 UP000054653 UP000005408
UP000235965 UP000069272 UP000053240 UP000069940 UP000249989 UP000000673 UP000075880 UP000075883 UP000075900 UP000075902 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000027135 UP000030765 UP000075885 UP000075881 UP000007151 UP000075920 UP000076408 UP000075886 UP000092462 UP000002320 UP000183832 UP000075884 UP000076858 UP000015103 UP000007819 UP000000305 UP000283509 UP000053454 UP000075901 UP000094527 UP000198287 UP000030742 UP000030746 UP000009046 UP000019118 UP000095280 UP000215902 UP000054359 UP000245119 UP000054653 UP000005408
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
Q1HQ47
Q2F682
S4PG03
C5IU79
A0A437BJH0
A0A194PJ03
+ More
A0A2A4J839 A0A2H1WMR5 A0A0L7KS43 A0A336L332 A0A1L8E0T4 D6WMM9 U5EUN0 A0A2Z5RE44 A0A2J7QTH1 A0A182FQB7 A0A194RCL7 A0A182GWF1 A0A1Q3G3V0 W5JVS7 A0A0P6IWM1 A0A2M4AET7 A0A1B6DUK8 T1DNG1 A0A2M3ZH85 A0A2M4BN00 A0A182IQW4 A0A182MDB5 A0A182R385 A0A182TNU5 A0A182VLM9 A0A182XE67 A0A182LCC4 Q7PQQ3 A0A182HJ56 A0A067R707 A0A084VT63 A0A182PAH4 A0A182K1D2 A0A212EP23 A0A1B6H9E0 A0A182VXG5 A0A170ZT89 A0A182XWU0 A0A182QLM4 A0A1B0GPW4 A0A1B6L4T4 B0W8J6 R4WRJ5 A0A1Q3FH56 A0A1J1J8G1 A0A023ERF4 A0A182NFS3 A0A0P6GDX1 A0A164X3A6 T1HWW5 A0A0P5M0M1 V5G6Y6 A0A0P6IAQ5 A0A2S2P120 A0A0N7Z8U3 J9K5R3 A0A0C5Q4W6 E9GAE4 A0A069DU83 A0A3R7SZS6 A0A0P6CFA3 A0A023F9T3 A0A2P1JJ64 A0A224XDB6 A0A2H8TLD4 A0A1Y1KBI4 A0A0L8H5J4 R4G2N3 A0A0V0GA37 A0A182SBQ9 D3PJB8 A0A0P5G6F5 A0A1D2MS27 A0A226ELI4 A0A2L2XYP2 A0A0P5ZHN6 U4UK34 J3JWC0 V3ZNV5 A0A0N8CY74 E0VCW6 N6U0Y1 A0A1I8JHX3 A0A087TI66 A0A1I8IEN0 T1JA51 A0A1I8IA73 A0A2T7PFS0 A0A0V1DFM4 A0A267E1V2 K1RZE2 A0A0V1DGP9 A0A023GKA1 I1VYX6
A0A2A4J839 A0A2H1WMR5 A0A0L7KS43 A0A336L332 A0A1L8E0T4 D6WMM9 U5EUN0 A0A2Z5RE44 A0A2J7QTH1 A0A182FQB7 A0A194RCL7 A0A182GWF1 A0A1Q3G3V0 W5JVS7 A0A0P6IWM1 A0A2M4AET7 A0A1B6DUK8 T1DNG1 A0A2M3ZH85 A0A2M4BN00 A0A182IQW4 A0A182MDB5 A0A182R385 A0A182TNU5 A0A182VLM9 A0A182XE67 A0A182LCC4 Q7PQQ3 A0A182HJ56 A0A067R707 A0A084VT63 A0A182PAH4 A0A182K1D2 A0A212EP23 A0A1B6H9E0 A0A182VXG5 A0A170ZT89 A0A182XWU0 A0A182QLM4 A0A1B0GPW4 A0A1B6L4T4 B0W8J6 R4WRJ5 A0A1Q3FH56 A0A1J1J8G1 A0A023ERF4 A0A182NFS3 A0A0P6GDX1 A0A164X3A6 T1HWW5 A0A0P5M0M1 V5G6Y6 A0A0P6IAQ5 A0A2S2P120 A0A0N7Z8U3 J9K5R3 A0A0C5Q4W6 E9GAE4 A0A069DU83 A0A3R7SZS6 A0A0P6CFA3 A0A023F9T3 A0A2P1JJ64 A0A224XDB6 A0A2H8TLD4 A0A1Y1KBI4 A0A0L8H5J4 R4G2N3 A0A0V0GA37 A0A182SBQ9 D3PJB8 A0A0P5G6F5 A0A1D2MS27 A0A226ELI4 A0A2L2XYP2 A0A0P5ZHN6 U4UK34 J3JWC0 V3ZNV5 A0A0N8CY74 E0VCW6 N6U0Y1 A0A1I8JHX3 A0A087TI66 A0A1I8IEN0 T1JA51 A0A1I8IA73 A0A2T7PFS0 A0A0V1DFM4 A0A267E1V2 K1RZE2 A0A0V1DGP9 A0A023GKA1 I1VYX6
PDB
1LWD
E-value=6.14212e-178,
Score=1603
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00480 Glutathione metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
00480 Glutathione metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
GO
PANTHER
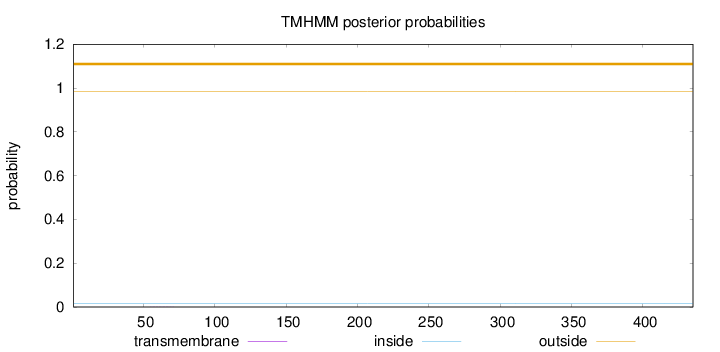
Topology
Length:
435
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00505999999999999
Exp number, first 60 AAs:
0.00119
Total prob of N-in:
0.01496
outside
1 - 435
Population Genetic Test Statistics
Pi
205.507663
Theta
176.522733
Tajima's D
0.61056
CLR
0.656967
CSRT
0.545372731363432
Interpretation
Uncertain
