Pre Gene Modal
BGIBMGA007501
Annotation
PREDICTED:_O-glucosyltransferase_rumi_homolog_[Papilio_machaon]
Full name
O-glucosyltransferase rumi homolog
+ More
O-glucosyltransferase rumi
O-glucosyltransferase rumi
Location in the cell
Cytoplasmic Reliability : 1.388 Nuclear Reliability : 1.302
Sequence
CDS
ATGTTAGATAGACCGTCGCTTATTATCTTATATTTTGTTTTATGTACTATTAGCTGCTATTTTTGTGAACATTATTGCAGCGGCGCCGAAAACACGTGTAAAGAAGATAAAAATAAATATAGTAGAGATGTAAATGAGTGGACTTCTAACTTAAAGAATGAAATAAAACGAGCCTTAGAACAATATGCTCCTTGCAAGAAAGAAAATTGTAGCTGTCATAGATCAGTAATTGATAGTGATCTGGCTCCATACCAGGATGGTATAACCAATGAAATGTTCATCTCAGCTCGAGCAAAGGGTACCAAGTACCAGGTCATAGATGGCAAGTTGTACCGAGAGGAAAATTGCCATTTTCCAGCCAGGTGTGCTGGTGTTGAACATTATATGAAATCTTTGAATTTAAAGAAGCTCTCAAATATGGAAGTATCCATTAATACTAGAGATTGGCCACAAGTTAACAAACTTTGGGGCCAAAAAAAAGCACCAGTGCTCTCTTTTAGTAAGACACAAGACTATTATGATATTATGTATCCTGCTTGGAGCTTTTGGGAAGGAGGGCCAGCCATAGCACTATACCCCACGGGTATAGGACGATGGGATGAACATAGAAAATCAATTTCCCTTGCAGCCAAAAAGAGCCCTTGGGATAAGAAAAAAGCAAAAGCATTCTTCCGAGGTTCAAGAACTAGTGAAGAAAGAGATGCTTTAATATTACTATCTCGTTCTAAACCTGATCTTGTAGATGCACAATATACCAAGAATCAGGCTTGGAAGTCAGATGCAGATACACTACATGCTCCTGCCGCCTCAGAAGTGTCCTTTGAAGATCATTGCAAATATAAATATCTGTTTAACTACAGAGGCGTAGCAGCTAGTTTCAGATTTAAACATTTATTTCTATGCAAATCCTTGGTTTTTCATGTCGGTGATGAATGGCTAGAATTCTTCTATCCGTCACTGAAGCCTTGGGTGCATTATATACCAATTAGCCCAAAGGCAAGCAAAGATGAAATTCAACAAGTCATAGAATTCTTTAAAGCTGAGGATGCACTTGCAAAGGAAATAGCAGATAGGGGATTTCAACATATTTGGGAAAACTTAGATGATGATGATGTGAAATGTTATTGGCAAATATTATTGAAGAAATATTCTAAACTAATAAAATATAAAGTTGTAAAAGATAAAGATCTGATAGAAATATAA
Protein
MLDRPSLIILYFVLCTISCYFCEHYCSGAENTCKEDKNKYSRDVNEWTSNLKNEIKRALEQYAPCKKENCSCHRSVIDSDLAPYQDGITNEMFISARAKGTKYQVIDGKLYREENCHFPARCAGVEHYMKSLNLKKLSNMEVSINTRDWPQVNKLWGQKKAPVLSFSKTQDYYDIMYPAWSFWEGGPAIALYPTGIGRWDEHRKSISLAAKKSPWDKKKAKAFFRGSRTSEERDALILLSRSKPDLVDAQYTKNQAWKSDADTLHAPAASEVSFEDHCKYKYLFNYRGVAASFRFKHLFLCKSLVFHVGDEWLEFFYPSLKPWVHYIPISPKASKDEIQQVIEFFKAEDALAKEIADRGFQHIWENLDDDDVKCYWQILLKKYSKLIKYKVVKDKDLIEI
Summary
Description
Protein O-glucosyltransferase. Catalyzes the reaction that attaches glucose through an O-glycosidic linkage to a conserved serine residue found in the consensus sequence C-X-S-X-[PA]-C in epidermal growth factor-like repeats. Regulates Notch signaling by glucosylating Notch in the ER, glucosylation is required for the correct folding and cleavage of Notch.
Protein O-glucosyltransferase. Catalyzes the reaction that attaches glucose through an O-glycosidic linkage to a conserved serine residue found in the consensus sequence C-X-S-X-[PA]-C in epidermal growth factor-like repeats (PubMed:27428513). Regulates Notch signaling by glucosylating Notch in the ER, glucosylation is required for the correct folding and cleavage of Notch.
Protein O-glucosyltransferase. Catalyzes the reaction that attaches glucose through an O-glycosidic linkage to a conserved serine residue found in the consensus sequence C-X-S-X-[PA]-C in epidermal growth factor-like repeats (PubMed:27428513). Regulates Notch signaling by glucosylating Notch in the ER, glucosylation is required for the correct folding and cleavage of Notch.
Similarity
Belongs to the glycosyltransferase 90 family.
Keywords
Complete proteome
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Reference proteome
Secreted
Signal
Transferase
Notch signaling pathway
3D-structure
Feature
chain O-glucosyltransferase rumi homolog
Uniprot
A0A2H1WMN1
H9JDA4
A0A2A4J764
A0A3S2NGL2
A0A194R8S8
A0A194PQJ1
+ More
A0A182H7G6 A0A182XX24 A0A1B0CVG1 A0A182JUK6 A0A1Q3EX11 A0A182QC01 W5JGS3 A0A2M4BQH3 A0A2M4BQH5 D6WKX5 A0A182NCX2 A0A336LJ21 A0A2M4BQH7 A0A182PS68 Q16QY8 A0A182RX70 A0A182FPF5 A0A067R6Q8 B0X1Q4 A0A182JFZ3 A0A182M506 A0A182W175 A0NDG6 A0A1W4XG66 A0A182VID6 A0A182L706 A0A182XG15 A0A182U7P6 A0A0N7ZA70 E9GPM7 A0A182ID61 Q29AU6 A0A151XD45 E9IUB8 B4G5X6 A0A232EDB9 A0A164KP06 A0A0P5TNA4 A0A195FGP7 B3LZQ3 A0A0P6AAJ7 A0A0P5M9X3 A0A0P6IJ49 A0A1J1IR89 A0A0P5XKM7 A0A0P5A993 A0A158NGW7 F4WZU1 B4R0R3 A0A195B6N0 A0A1A9Z9Z7 A0A1W4VK38 B3P8J6 A0A226E664 B4PMF4 B4HET9 A0A0L0BN71 Q8T045 A0A0P6FRM3 A0A0P6F3Q6 A0A151II91 W8BTG6 A0A1B6HQ52 A0A0K8WM16 E2ARY7 A0A1A9W1A1 K1Q9M3 A0A3B0KBF9 A0A0P5LZ81 E0W3T1 A0A293LAX1 A0A099ZT40 A0A1B6DS49 A0A093HRN4 A0A091HKU8 A0A195DWF1 A0A093EZX5 A0A091U372 A0A093P427 A0A091K2M9 K7J7V2 A0A0A0ARU6 A0A091IZJ3 H0ZTH7 A0A091ELJ1 B4KBE5 A0A093F916 A0A091GH44 A0A2Y9DGJ0 A0A0Q3M9I3 U3JFL6 A0A093G990 A0A1U7QG35 A0A091U4A7
A0A182H7G6 A0A182XX24 A0A1B0CVG1 A0A182JUK6 A0A1Q3EX11 A0A182QC01 W5JGS3 A0A2M4BQH3 A0A2M4BQH5 D6WKX5 A0A182NCX2 A0A336LJ21 A0A2M4BQH7 A0A182PS68 Q16QY8 A0A182RX70 A0A182FPF5 A0A067R6Q8 B0X1Q4 A0A182JFZ3 A0A182M506 A0A182W175 A0NDG6 A0A1W4XG66 A0A182VID6 A0A182L706 A0A182XG15 A0A182U7P6 A0A0N7ZA70 E9GPM7 A0A182ID61 Q29AU6 A0A151XD45 E9IUB8 B4G5X6 A0A232EDB9 A0A164KP06 A0A0P5TNA4 A0A195FGP7 B3LZQ3 A0A0P6AAJ7 A0A0P5M9X3 A0A0P6IJ49 A0A1J1IR89 A0A0P5XKM7 A0A0P5A993 A0A158NGW7 F4WZU1 B4R0R3 A0A195B6N0 A0A1A9Z9Z7 A0A1W4VK38 B3P8J6 A0A226E664 B4PMF4 B4HET9 A0A0L0BN71 Q8T045 A0A0P6FRM3 A0A0P6F3Q6 A0A151II91 W8BTG6 A0A1B6HQ52 A0A0K8WM16 E2ARY7 A0A1A9W1A1 K1Q9M3 A0A3B0KBF9 A0A0P5LZ81 E0W3T1 A0A293LAX1 A0A099ZT40 A0A1B6DS49 A0A093HRN4 A0A091HKU8 A0A195DWF1 A0A093EZX5 A0A091U372 A0A093P427 A0A091K2M9 K7J7V2 A0A0A0ARU6 A0A091IZJ3 H0ZTH7 A0A091ELJ1 B4KBE5 A0A093F916 A0A091GH44 A0A2Y9DGJ0 A0A0Q3M9I3 U3JFL6 A0A093G990 A0A1U7QG35 A0A091U4A7
EC Number
2.4.1.-
Pubmed
19121390
26354079
26483478
25244985
20920257
23761445
+ More
18362917 19820115 17510324 24845553 12364791 20966253 21292972 15632085 21282665 17994087 28648823 21347285 21719571 17550304 26108605 10731132 12537572 12537569 18243100 27807076 27428513 24495485 20798317 22992520 20566863 20075255 20360741
18362917 19820115 17510324 24845553 12364791 20966253 21292972 15632085 21282665 17994087 28648823 21347285 21719571 17550304 26108605 10731132 12537572 12537569 18243100 27807076 27428513 24495485 20798317 22992520 20566863 20075255 20360741
EMBL
ODYU01009673
SOQ54267.1
BABH01030171
NWSH01002582
PCG67925.1
RSAL01000046
+ More
RVE50548.1 KQ460615 KPJ13655.1 KQ459602 KPI93400.1 JXUM01116197 KQ565958 KXJ70493.1 AJWK01030488 GFDL01015202 JAV19843.1 AXCN02001559 ADMH02001267 ETN63286.1 GGFJ01006194 MBW55335.1 GGFJ01006195 MBW55336.1 KQ971343 EFA03548.1 UFQT01000012 SSX17605.1 GGFJ01006196 MBW55337.1 CH477727 EAT36834.1 KK852661 KDR19100.1 DS232266 EDS38782.1 AXCM01004697 AAAB01008880 EAU76986.1 GDRN01103765 JAI58024.1 GL732557 EFX78410.1 APCN01005034 CM000070 EAL27253.1 KQ982294 KYQ58269.1 GL765937 EFZ15836.1 CH479179 EDW23731.1 NNAY01006883 OXU16351.1 LRGB01003275 KZS03426.1 GDIP01124302 JAL79412.1 KQ981625 KYN39174.1 CH902617 EDV43047.1 GDIP01033296 JAM70419.1 GDIQ01162930 JAK88795.1 GDIQ01010207 JAN84530.1 CVRI01000057 CRL02068.1 GDIP01070706 JAM33009.1 GDIP01202339 JAJ21063.1 ADTU01015296 GL888480 EGI60366.1 CM000364 EDX13986.1 KQ976574 KYM80173.1 CH954182 EDV54091.1 LNIX01000006 OXA53103.1 CM000160 EDW98059.1 CH480815 EDW43250.1 JRES01001623 KNC21462.1 AE014297 AY069564 AAL39709.1 AAN13920.1 GDIQ01046377 JAN48360.1 GDIQ01062015 JAN32722.1 KQ977492 KYN02298.1 GAMC01004078 JAC02478.1 GECU01030904 JAS76802.1 GDHF01000215 JAI52099.1 GL442209 EFN63761.1 JH816276 EKC30658.1 OUUW01000007 SPP83006.1 GDIQ01162929 JAK88796.1 AAZO01007449 DS235882 EEB20287.1 GFWV01000188 MAA24918.1 KL897324 KGL84343.1 GEDC01008829 JAS28469.1 KL206729 KFV85323.1 KL217544 KFO96486.1 KQ980204 KYN17218.1 KK379424 KFV47561.1 KK497091 KFQ66178.1 KL225051 KFW67027.1 KK539516 KFP30453.1 AAZX01012650 KL872973 KGL97294.1 KK501141 KFP12943.1 ABQF01024500 KK718540 KFO57462.1 CH933806 EDW16873.1 KK625205 KFV53060.1 KL447422 KFO73457.1 LMAW01002593 KQK79357.1 AGTO01011023 KL215807 KFV66770.1 KK414333 KFQ84872.1
RVE50548.1 KQ460615 KPJ13655.1 KQ459602 KPI93400.1 JXUM01116197 KQ565958 KXJ70493.1 AJWK01030488 GFDL01015202 JAV19843.1 AXCN02001559 ADMH02001267 ETN63286.1 GGFJ01006194 MBW55335.1 GGFJ01006195 MBW55336.1 KQ971343 EFA03548.1 UFQT01000012 SSX17605.1 GGFJ01006196 MBW55337.1 CH477727 EAT36834.1 KK852661 KDR19100.1 DS232266 EDS38782.1 AXCM01004697 AAAB01008880 EAU76986.1 GDRN01103765 JAI58024.1 GL732557 EFX78410.1 APCN01005034 CM000070 EAL27253.1 KQ982294 KYQ58269.1 GL765937 EFZ15836.1 CH479179 EDW23731.1 NNAY01006883 OXU16351.1 LRGB01003275 KZS03426.1 GDIP01124302 JAL79412.1 KQ981625 KYN39174.1 CH902617 EDV43047.1 GDIP01033296 JAM70419.1 GDIQ01162930 JAK88795.1 GDIQ01010207 JAN84530.1 CVRI01000057 CRL02068.1 GDIP01070706 JAM33009.1 GDIP01202339 JAJ21063.1 ADTU01015296 GL888480 EGI60366.1 CM000364 EDX13986.1 KQ976574 KYM80173.1 CH954182 EDV54091.1 LNIX01000006 OXA53103.1 CM000160 EDW98059.1 CH480815 EDW43250.1 JRES01001623 KNC21462.1 AE014297 AY069564 AAL39709.1 AAN13920.1 GDIQ01046377 JAN48360.1 GDIQ01062015 JAN32722.1 KQ977492 KYN02298.1 GAMC01004078 JAC02478.1 GECU01030904 JAS76802.1 GDHF01000215 JAI52099.1 GL442209 EFN63761.1 JH816276 EKC30658.1 OUUW01000007 SPP83006.1 GDIQ01162929 JAK88796.1 AAZO01007449 DS235882 EEB20287.1 GFWV01000188 MAA24918.1 KL897324 KGL84343.1 GEDC01008829 JAS28469.1 KL206729 KFV85323.1 KL217544 KFO96486.1 KQ980204 KYN17218.1 KK379424 KFV47561.1 KK497091 KFQ66178.1 KL225051 KFW67027.1 KK539516 KFP30453.1 AAZX01012650 KL872973 KGL97294.1 KK501141 KFP12943.1 ABQF01024500 KK718540 KFO57462.1 CH933806 EDW16873.1 KK625205 KFV53060.1 KL447422 KFO73457.1 LMAW01002593 KQK79357.1 AGTO01011023 KL215807 KFV66770.1 KK414333 KFQ84872.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000069940
+ More
UP000249989 UP000076408 UP000092461 UP000075881 UP000075886 UP000000673 UP000007266 UP000075884 UP000075885 UP000008820 UP000075900 UP000069272 UP000027135 UP000002320 UP000075880 UP000075883 UP000075920 UP000007062 UP000192223 UP000075903 UP000075882 UP000076407 UP000075902 UP000000305 UP000075840 UP000001819 UP000075809 UP000008744 UP000215335 UP000076858 UP000078541 UP000007801 UP000183832 UP000005205 UP000007755 UP000000304 UP000078540 UP000092445 UP000192221 UP000008711 UP000198287 UP000002282 UP000001292 UP000037069 UP000000803 UP000078542 UP000000311 UP000091820 UP000005408 UP000268350 UP000009046 UP000053641 UP000053584 UP000054308 UP000078492 UP000054081 UP000002358 UP000053858 UP000053119 UP000007754 UP000052976 UP000009192 UP000053760 UP000248480 UP000051836 UP000016665 UP000053875 UP000189706
UP000249989 UP000076408 UP000092461 UP000075881 UP000075886 UP000000673 UP000007266 UP000075884 UP000075885 UP000008820 UP000075900 UP000069272 UP000027135 UP000002320 UP000075880 UP000075883 UP000075920 UP000007062 UP000192223 UP000075903 UP000075882 UP000076407 UP000075902 UP000000305 UP000075840 UP000001819 UP000075809 UP000008744 UP000215335 UP000076858 UP000078541 UP000007801 UP000183832 UP000005205 UP000007755 UP000000304 UP000078540 UP000092445 UP000192221 UP000008711 UP000198287 UP000002282 UP000001292 UP000037069 UP000000803 UP000078542 UP000000311 UP000091820 UP000005408 UP000268350 UP000009046 UP000053641 UP000053584 UP000054308 UP000078492 UP000054081 UP000002358 UP000053858 UP000053119 UP000007754 UP000052976 UP000009192 UP000053760 UP000248480 UP000051836 UP000016665 UP000053875 UP000189706
Interpro
SUPFAM
SSF55856
SSF55856
Gene 3D
ProteinModelPortal
A0A2H1WMN1
H9JDA4
A0A2A4J764
A0A3S2NGL2
A0A194R8S8
A0A194PQJ1
+ More
A0A182H7G6 A0A182XX24 A0A1B0CVG1 A0A182JUK6 A0A1Q3EX11 A0A182QC01 W5JGS3 A0A2M4BQH3 A0A2M4BQH5 D6WKX5 A0A182NCX2 A0A336LJ21 A0A2M4BQH7 A0A182PS68 Q16QY8 A0A182RX70 A0A182FPF5 A0A067R6Q8 B0X1Q4 A0A182JFZ3 A0A182M506 A0A182W175 A0NDG6 A0A1W4XG66 A0A182VID6 A0A182L706 A0A182XG15 A0A182U7P6 A0A0N7ZA70 E9GPM7 A0A182ID61 Q29AU6 A0A151XD45 E9IUB8 B4G5X6 A0A232EDB9 A0A164KP06 A0A0P5TNA4 A0A195FGP7 B3LZQ3 A0A0P6AAJ7 A0A0P5M9X3 A0A0P6IJ49 A0A1J1IR89 A0A0P5XKM7 A0A0P5A993 A0A158NGW7 F4WZU1 B4R0R3 A0A195B6N0 A0A1A9Z9Z7 A0A1W4VK38 B3P8J6 A0A226E664 B4PMF4 B4HET9 A0A0L0BN71 Q8T045 A0A0P6FRM3 A0A0P6F3Q6 A0A151II91 W8BTG6 A0A1B6HQ52 A0A0K8WM16 E2ARY7 A0A1A9W1A1 K1Q9M3 A0A3B0KBF9 A0A0P5LZ81 E0W3T1 A0A293LAX1 A0A099ZT40 A0A1B6DS49 A0A093HRN4 A0A091HKU8 A0A195DWF1 A0A093EZX5 A0A091U372 A0A093P427 A0A091K2M9 K7J7V2 A0A0A0ARU6 A0A091IZJ3 H0ZTH7 A0A091ELJ1 B4KBE5 A0A093F916 A0A091GH44 A0A2Y9DGJ0 A0A0Q3M9I3 U3JFL6 A0A093G990 A0A1U7QG35 A0A091U4A7
A0A182H7G6 A0A182XX24 A0A1B0CVG1 A0A182JUK6 A0A1Q3EX11 A0A182QC01 W5JGS3 A0A2M4BQH3 A0A2M4BQH5 D6WKX5 A0A182NCX2 A0A336LJ21 A0A2M4BQH7 A0A182PS68 Q16QY8 A0A182RX70 A0A182FPF5 A0A067R6Q8 B0X1Q4 A0A182JFZ3 A0A182M506 A0A182W175 A0NDG6 A0A1W4XG66 A0A182VID6 A0A182L706 A0A182XG15 A0A182U7P6 A0A0N7ZA70 E9GPM7 A0A182ID61 Q29AU6 A0A151XD45 E9IUB8 B4G5X6 A0A232EDB9 A0A164KP06 A0A0P5TNA4 A0A195FGP7 B3LZQ3 A0A0P6AAJ7 A0A0P5M9X3 A0A0P6IJ49 A0A1J1IR89 A0A0P5XKM7 A0A0P5A993 A0A158NGW7 F4WZU1 B4R0R3 A0A195B6N0 A0A1A9Z9Z7 A0A1W4VK38 B3P8J6 A0A226E664 B4PMF4 B4HET9 A0A0L0BN71 Q8T045 A0A0P6FRM3 A0A0P6F3Q6 A0A151II91 W8BTG6 A0A1B6HQ52 A0A0K8WM16 E2ARY7 A0A1A9W1A1 K1Q9M3 A0A3B0KBF9 A0A0P5LZ81 E0W3T1 A0A293LAX1 A0A099ZT40 A0A1B6DS49 A0A093HRN4 A0A091HKU8 A0A195DWF1 A0A093EZX5 A0A091U372 A0A093P427 A0A091K2M9 K7J7V2 A0A0A0ARU6 A0A091IZJ3 H0ZTH7 A0A091ELJ1 B4KBE5 A0A093F916 A0A091GH44 A0A2Y9DGJ0 A0A0Q3M9I3 U3JFL6 A0A093G990 A0A1U7QG35 A0A091U4A7
PDB
5F87
E-value=1.27808e-112,
Score=1039
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum lumen
Secreted
Secreted
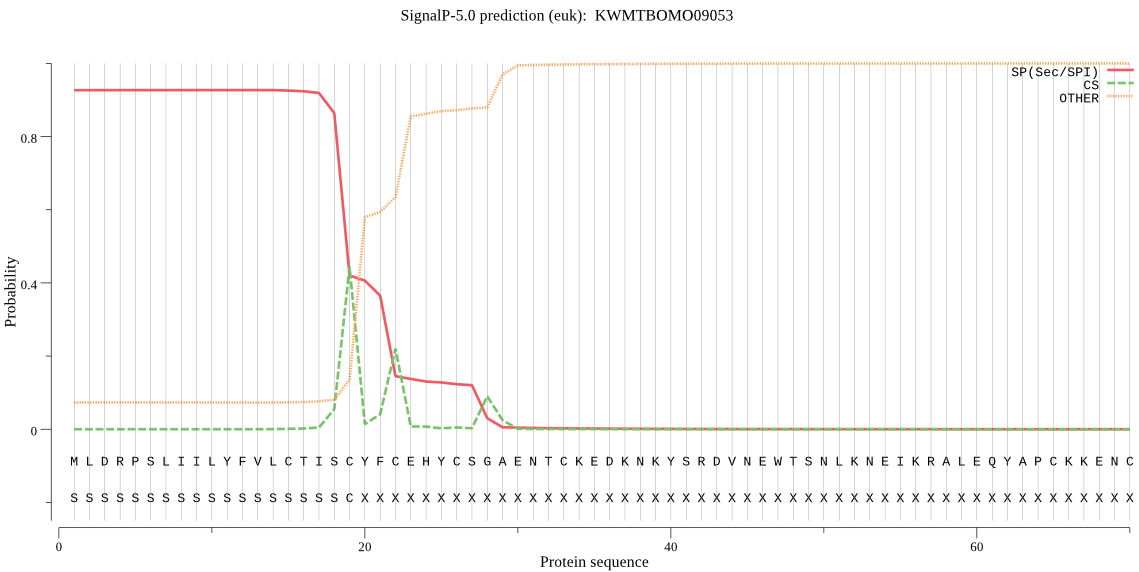
SignalP
Position: 1 - 19,
Likelihood: 0.927324
Length:
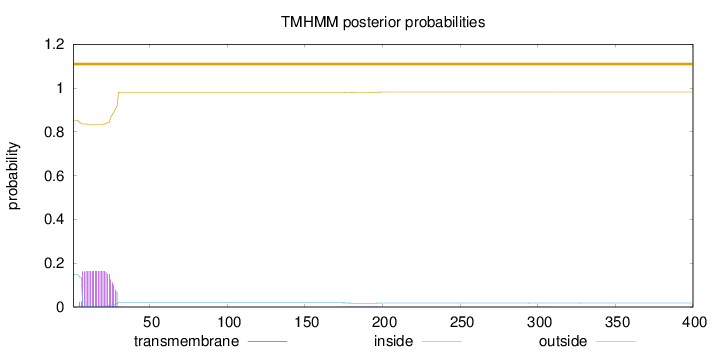
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.50371
Exp number, first 60 AAs:
3.45324
Total prob of N-in:
0.14698
outside
1 - 400
Population Genetic Test Statistics
Pi
223.664157
Theta
176.131704
Tajima's D
0.859654
CLR
1.613191
CSRT
0.620418979051047
Interpretation
Uncertain
